Abstract
The retroviral protease is a key enzyme in a viral multienzyme complex that initiates an ordered sequence of events leading to virus assembly and propagation. Viral peptides are initially synthesized as polyprotein precursors; these precursors undergo a number of proteolytic cleavages executed by the protease in a specific and presumably ordered manner. To determine the role of individual protease cleavage sites in Ty1, a retrotransposon from Saccharomyces cerevisiae, the cleavage sites were systematically mutagenized. Altering the cleavage sites of the yeast Ty1 retrotransposon produces mutants with distinct retrotransposition phenotypes. Blocking the Gag/PR site also blocks cleavage at the other two cleavage sites, PR/IN and IN/RT. In contrast, mutational block of the PR/IN or IN/RT sites does not prevent cleavage at the other two sites. Retrotransposons with mutations in each of these sites have transposition defects. Mutations in the PR/IN and IN/RT sites, but not in the Gag/PR site, can be complemented in trans by endogenous Ty1 copies. Hence, the digestion of the Gag/PR site and release of the protease N terminus is a prerequisite for processing at the remaining sites; cleavage of PR/IN is not required for the cleavage of IN/RT, and vice versa. Of the three cleavage sites in the Gag-Pol precursor, the Gag/PR site is processed first. Thus, Ty1 Gag-Pol processing proceeds by an ordered pathway.
The transposition process of the Ty1 retrotransposon from the yeast Saccharomyces cerevisiae shares numerous similarities with typical retroviral life cycles (1, 2, 29). Ty1 in its DNA form is integrated in the host genome (8), and the transcription of Ty1 yields an approximately 5.5-kb-long mRNA which contains a +1 frameshift signal (9, 14, 19); translation of this Ty1 mRNA yields two polyprotein precursors, a Gag precursor of 49 kDa and a 199-kDa Gag-Pol precursor.
We and others have previously named Ty1 proteins based on their apparent molecular weight as judged from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Several Ty1-encoded proteins do not migrate on gels according to their predicted molecular weight. As the masses of all Ty1 peptides is now known, we are revising our Ty1 protein nomenclature to reflect this and also to conform with retroviral standards (Table 1).
TABLE 1.
Ty1-encoded protein nomenclature
| Systematic namea | Old name(s) | Suggested name(s) |
|---|---|---|
| Gag (primary translation product) | Gag-p58, p1 | Gag-p49 |
| Gag-Pol (primary translation product) | Gag-Pol-p190 | Gag-Pol-p199 |
| CA (capsid) | Gag-p54, p2 | CA, Gag-p45 |
| None assigned | Gag-p4 | Gag-p4 |
| PR (protease) | PR, p23 | PR, Pol-p20 |
| IN (integrase) | p90, p84 | IN, Pol-p71 |
| RT (reverse transcriptase) | p60 | RT, Pol-p63 |
From reference 24.
The primary translation product Gag-p49 is cleaved into two products, the 45-kDa CA protein, which assembles into virus-like particles (VLPs) (16) and is required for transposition (5), and a 4-kDa C-terminal peptide, Gag-p4, which is not required for transposition and whose existence is inferred from mutational analyses (30). Gag-p4 has not yet been directly detected in VLPs or cell extracts. The Gag-Pol-p199 protein also undergoes proteolytic processing and is cleaved into four proteins, CA, PR, IN, and RT (17, 28). The cleavage site in the Gag precursor has been precisely mapped by systematic mutagenesis and by C-terminal sequencing of mature Gag (30) and has been confirmed by mass spectroscopic analysis of VLP proteins (J. F. Lawler, Jr., Rick Newitt, Rudi Aebersold, and J. D. Boeke, unpublished data). The inferred Gag/p4 cleavage site is at the same position as the Gag/PR site in the Gag-Pol precursor determined by the N-terminal sequencing of mature PR produced by autoprocessing in Escherichia coli (Fig. 1) (J. F. Lawler, G. V. Merkulov, and J. D. Boeke, submitted for publication). For simplicity, we will refer to the Gag/p4 and Gag/PR cleavage site(s) collectively as the Gag/PR site. The PR/IN and IN/RT cleavage sites were previously determined by N-terminal sequencing of IN and RT, correspondingly (6, 32; G. Sharon and David Garfinkel, personal communication). Although Ty1 PR cleavage sites reveal little sequence similarity, their hydrophobicity profiles are similar (30), suggesting that hydrophobicity patterns (perhaps in combination with accessibility in the folded structure) rather than the primary sequences of the cleavage sites are recognized by the enzyme. This hypothesis is consistent with theories explaining the specificity of retroviral proteases (21, 34). Mutations near the Ty1 PR active site block processing and transposition as well, confirming the key role played by Ty1 PR (31, 39). A mutant in which the Gag/PR cleavage is blocked exhibits the same phenotypes as a PR active site mutant, suggesting that the cleavage of this site and release of the protease N terminus are required for the processing of the other two sites and for retrotransposition (30).
FIG. 1.
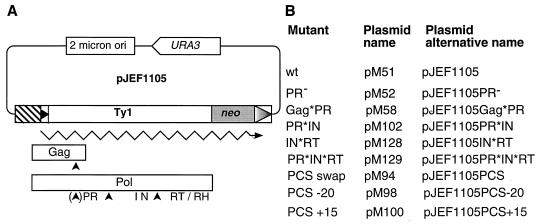
Plasmids and mutants. (A) Genetic map of the parental plasmid, pJEF1105 (4). Hatched box, GAL1 promoter; boxed triangles, LTR sequences; Gag and Pol, Ty1 primary translation products; neo, Ty1 marker gene; URA3, vector selectable marker; 2 micron ori, yeast 2 μm plasmid origin of replication; PR, IN, and RT/RH, regions of sequence similarity to retroviral proteins; arrowheads, PR cleavage sites. Plasmid backbones are not drawn to scale. (B) mutants and plasmids. wt, wild type.
In this study, the other two Ty1 cleavage sites were mutated both to confirm the locations of the cleavage sites inferred from protein sequencing (although certain mutations distant from cleavage sites can affect processing) and to study their roles in transposition. Substitutions or deletions in the PR/IN or IN/RT cleavage site or both sites inhibit Ty1 transposition but block proteolysis only at the affected site, suggesting that these two sites are functionally distinct from the critical Gag/PR site. Surprisingly, transposition of PR/IN and IN/RT cleavage site mutants is fully blocked only in host strains that fail to express endogenous Ty1 transposons (spt3 strains), not in strains that do (SPT3+ strains). The PR/IN and IN/RT site mutants therefore behave differently from the Gag/PR site mutant. While the processing of the Gag/PR cleavage site is a prerequisite for the processing of the other two, cleavage of PR/IN or IN/RT is not required for the processing to be completed. Systematic mutagenesis of the PR/IN and IN/RT cleavage sites reveals their essential role in the Ty1 life cycle and provides evidence for an ordered pathway of proteolytic cleavage in Ty1. Analysis of the biochemical defects in Ty1 transposition in the PR/IN, IN/RT, and PR/IN/RT cleavage site mutants showed that they are capable of forming VLPs and have RT activity; moreover, they make normal or near-normal levels of Ty1 cDNA. This suggests a late defect in transposition in these mutants, affecting either the integration reaction itself or transport or behavior of the preintegration complex.
The unique role of the Gag/PR cleavage site is specified by its position and not by its sequence. Relocation of the native Gag-PR cleavage site upstream or downstream of its native position rendered it inactive for cleavage and retrotransposition. However, replacement of the Gag/PR cleavage site with the PR/IN cleavage site resulted in a retrotransposon with normal proteolytic cleavage and retrotransposition phenotypes.
MATERIALS AND METHODS
Yeast strains.
The transposition assays were done using congenic yeast strains YH10 (MATa ura3-52 his4-539 lys2-801 GAL+) (38) and YH51 (MATa ura3-52 his4-539 lys2-801 spt3 GAL+). Cell extracts were prepared from YH51 strains carrying plasmids with mutant Gal-Ty1-neo constructs.
Vectors and plasmids.
All mutants were derived from on the pJEF1105 (pGAL-Ty1-neo) expression vector (Fig. 1) (4). The PR− mutant, pGM17, as well as the Gag*PR mutant (previously referred to as amino acid substitution mutant s3 at the Gag/PR cleavage site) were made as described earlier (30, 31). The PR cleavage site mutations were six-codon (AAGSAA) block substitutions as described previously (30).
To alter the PR/IN site, the Ty1 fragment extending from the 5′ long terminal repeat (LTR) XhoI site to the SalI site was subcloned into the pBluescript KS(+) vector (Stratagene); to change the IN/RT site, we inserted the Ty1 fragment between the SalI and BamHI sites into the same vector and performed site-directed mutagenesis on these plasmids as described by Kunkel (23). Oligonucleotide JB1204 (5′ TCAAATATCTCCGTACCCGCTGCTGGATCCGCTGCTACAAGTGAAAGTACACGC 3′) was used to change 18 nucleotides (bold; PR*IN mutant), and JB1205 (5′ TCGAAGAAACGAATTCACGCTGCTGGATCCGCTGCTGCAGTAAAATCAATCAA 3′) was used to make the IN*RT mutant. After mutants were identified by digestion with BamHI (site underlined), the Ty1 fragments between BstEII and KpnI (PR*IN mutant) or between KpnI and AflII (IN*RT mutant) sites were subcloned into pJEF1105. The PR*IN*RT mutant was constructed by subcloning a KpnI-AflII fragment of pJEF1105IN*RT into pJEF1105PR*IN. The PR cleavage site (PCS) swap mutant was constructed using the same strategy; oligonucleotide JB1201 (5′ AATTCGAAATCGAAAACAGGTACCATCAATAATGTACACACATCTAATAACTCTCCC 3′) was used to change 21 nucleotides (bold) in the PCS swap mutant; mutants were identified by digestion with KpnI (site underlined). This mutation results in a RAH/NVS→TIN/NVH change.
Immunoblotting.
Cultures were grown at 22°C in SC-Ura galactose medium. The starting cell density was 0.5 A600, and the cells were collected when the density reached 2 A600 (about 30 h at 22°C). Whole-cell extract samples were prepared for immunoblotting as described elsewhere (20) except that 2.5-ml samples of cells were used. Cell debris was pelleted by centrifugation for 3 min at 14,000 rpm, the supernatant was transferred to a fresh tube, and 10 μl of the supernatant, containing approximately 5 μg of protein, was mixed with an equal volume of 2× SDS-PAGE sample buffer, boiled, and loaded onto the gel.
Proteins were transferred onto Protran membranes (Schleicher & Schuell) in Tris-glycine buffer containing 20% methanol at 200 mA for 30 min. Membranes were blocked in 20 ml of phosphate-buffered saline (PBS) containing 5% milk, washed three times in PBS, and incubated with the indicated antibodies in PBS. After three subsequent washes in PBS, filters were incubated with the appropriate secondary antibodies, then incubated with ECL fluorescent reagent (Amersham), and exposed to X-ray film. Anti-Gag (anti-VLP; R2-F) and anti-IN (8B11) antibodies are described elsewhere (12, 31). For all procedures using the R2-F antibody, PBS containing 0.1% Tween-20 was substituted for PBS.
Transposition assay.
Yeast strain YH10 and YH51 cells were transformed to Ura+ (prototrophy) with plasmids carrying wild-type and mutant Gal-Ty1-neo retroelements. Transformants were patched onto SC-Ura glucose plates, grown at 30°C for 24 h, then replica plated onto SC-Ura galactose plates, and incubated at 22°C for 48 h to induce transposition. The patches were then replica plated onto YPD nonselective medium at 30°C overnight to allow loss of the donor plasmid. Cells that lost donor plasmid were selected by replica plating to SC–5-fluoro-orotic acid glucose medium and finally replica plated onto YPD medium containing G418 (75 μg/ml) to select for the cells that acquired genomic copies of Ty1-neo.
VLP isolation and cDNA analysis.
For VLP analysis, cell pellets from 500-ml cultures grown at 22°C were resuspended in 5 ml of buffer B-EDTA (12) and lysed with glass beads at 4°C. The extract was clarified by centrifugation at 17,000 rpm in a Sorvall SS-34 rotor for 10 min. Supernatant (5 ml) was loaded on a preformed linear 20 to 70% sucrose gradient in buffer B/EDTA and centrifuged for 18 h at 25,000 rpm at 4°C in a Beckman SW28 rotor. Gradients were fractionated, and RT activity associated with VLPs was determined as described elsewhere (12). For cDNA analysis, samples were processed as described elsewhere (Lawler et al., submitted).
RESULTS AND DISCUSSION
Mutagenesis of Ty1 PR cleavage sites in a GAL-Ty1-neo element.
The parental plasmid pJEF1105 was used to generate mutant Ty1-neo elements with block substitution mutations in PR cleavage sites (Fig. 1). pJEF1105 is a shuttle vector that contains both bacterial and yeast origins of replication and can be propagated in high copy number in bacteria and yeast. In these plasmids, a portion of the 5′ LTR of Ty1, containing the native Ty1 promoter, is replaced by the GAL1 promoter, so that the Ty1-neo element is driven by the GAL1 promoter and therefore transcribed at high levels upon induction with galactose. VLPs are then assembled within which Ty1-neo cDNA is synthesized; the cDNA is subsequently integrated into the genome by Ty1 IN. Cells containing the newly transposed Ty1-neo elements are easily identified by replica plating (Materials and Methods).
Primary Ty1 translation products and their subsequent proteolytic products are readily detected in extracts from cells carrying these plasmids (Fig. 2 and 3); defects in Ty1 processing are also easily detected using this expression system. The Gag/PR cleavage site was determined previously (30; Lawler et al., submitted); it was also shown that substitution of this site by the amino acid residues AAGSAA centered about the Gag/PR cleavage site blocked cleavage. The same approach was used to block the other two cleavage sites in Ty1 (Fig. 2). Similar results were obtained with 12-amino-acid (aa) deletion mutations centered about the three PR cleavage sites in our laboratory (data not shown) and with 2-aa deletion mutants studied by G. Sharon and D. J. Garfinkel (personal communication).
FIG. 2.
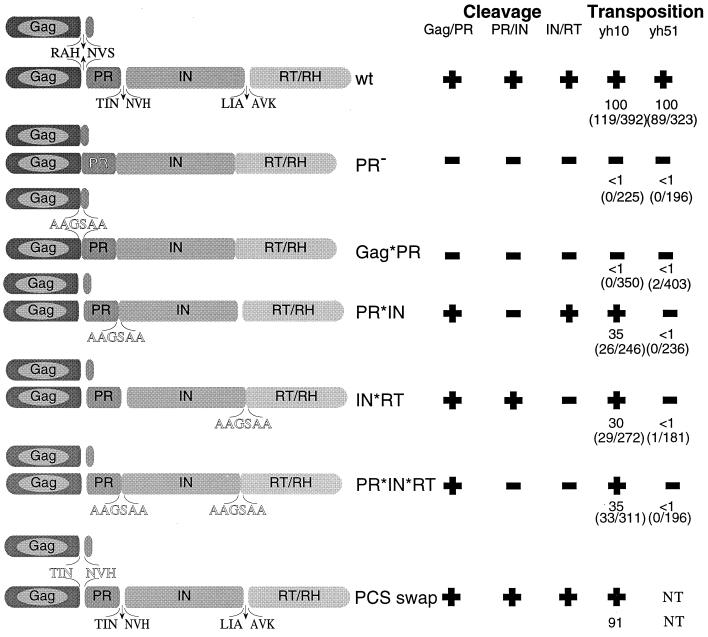
Mutations analyzed and summary of cleavage and transposition data. Shaded oblongs, Ty1 proteins; arrows, cleavage site locations. Mutant sequences are outlined. Transposition frequencies are indicated as percentage of wild type; raw data are in parentheses.
FIG. 3.
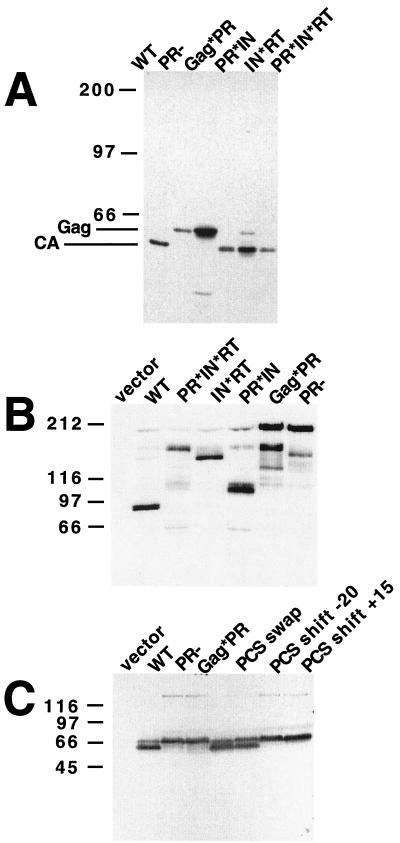
Immunoblots of Gag and Gag-Pol processing products. Cultures containing plasmids with mutations in the Ty1 PR cleavage sites were grown in liquid SC-Ura galactose medium at 22°C. Cells were pelleted, lysed, subjected to SDS-PAGE, transferred onto an Immobilon membrane, and incubated with anti-Gag (A and C) or anti-IN (B) antiserum. WT, wild type. Sizes are indicated in kilodaltons.
Mutations in Ty1 PR cleavage sites affect proteolytic cleavage differentially.
Ty1 Gag was not cleaved in cells expressing the mutant Ty1 with a substitution in the Gag/PR cleavage site (Gag*PR [Fig. 3A]). The Gag-Pol precursor is also largely uncut in the Gag*PR mutant. While most of the Gag-Pol-derived material observed in this mutant comigrates with the Gag-Pol-199 protein produced by the PR− (active site region) mutant, two additional, faster-moving bands are sometimes observed. We suspect that these smaller band result from proteolytic degradation during sample preparation because the amount of this material is very variable. The data suggest that cleavage between Gag and p4 in the 58-kDa Gag precursor and/or the cleavage site between Gag and PR in the 199-kDa Gag-Pol precursor is required for cleavage at the other sites. In other studies, we found that the N terminus of Ty1 PR produced by autoproteolysis of Gag-PR expressed in E. coli is NVSTS, indicating that this cleavage site defines the Ty1 PR N terminus (Lawler et al., submitted). The failure to observe proteolysis at any of the cleavage sites in the Gag*PR mutant could in principle be explained by one of two hypotheses: (i) cleavage of the N terminus is required for activity at the other sites, or (ii) the N-terminal residues which are altered in the Gag*PR mutant might be required for catalysis. The first possibility can be further split into two possible mechanisms: the N terminus may need to be liberated from CA to gain activity in trans, or a change in PR structure accompanies CA cleavage and activates it for cleavage at other sites. Possibility ii seems unlikely based on results from an earlier study (30), in which we constructed single amino acid substitution mutations altering residues 1 and 3 of Ty1 PR (mutants s3.4 and s3.6); these amino acid substitutions did not prevent catalysis. However, mutant s3.3, which blocked Gag/PR cleavage by mutating a residue in Gag, has the same phenotype as the Gag*PR block substitution mutant: failure to process Gag-Pol as well as Gag. Taken together, these results suggest that release of the protease N terminus is an essential step for its maturation and is required for subsequent processing of the other two sites.
In contrast, Ty1 Gag precursor was processed normally in cells expressing the PR*IN mutant, in which the cleavage site between PR and IN was altered by substitution. However, no mature IN was detected in these cells with anti-IN antibodies. Instead, a species with an apparent molecular mass of approximately 105 kDa was detected, as expected for cleavage at all sites except the PR/IN site (Fig. 3B). Hence, in the PR*IN mutant, the PR/IN site is effectively blocked by substitution, resulting in expression of a PR-IN fusion protein. We conclude that cleavage at the PR/IN site and subsequent release of the protease C terminus are not required for cleavage at the other sites. Thus, the cleavages at the protease N and C termini have different functional consequences. In a similar fashion, the human immunodeficiency virus type 1 (HIV-1) PR is activated by (auto)proteolytic cleavage at its N terminus (26).
Similarly, altering the cleavage site in the Ty1 IN*RT mutant did not hamper cleavage of the Gag precursor, as the mature Gag-p45 species was detected by immunoblot analysis (Fig. 3A). Anti-IN antiserum was used to detect a protein with an apparent molecular mass of approximately 145 kDa in this mutant, corresponding to the expected size of an IN-RT fusion protein (Fig. 3B). The Gag/PR and PR/IN sites were processed in the IN*RT mutant; thus, we conclude that blocking the IN/RT cleavage site did not affect cleavage at the other sites. Blocking both PR/IN and IN/RT sites in the PR*IN*RT double mutant allowed detection of a PR-IN-RT fusion protein with an apparent molecular mass of approximately 165 kDa with anti-IN antibodies; Gag was also processed normally in this mutant.
Systematic mutagenesis of PR cleavage sites performed on a number of retroviruses demonstrated that blocking the processing at some sites may inhibit cleavage at the others. Mutation of the NC/PR cleavage site of an avian retrovirus blocked cleavage at the other Gag sites (7). Mutations in the PR/RT cleavage site of HIV-1 inhibit processing at the downstream cleavage sites (25). Blocking the amino-terminal site of the HIV-1 PR did not block PR activity in vitro but significantly reduced infectivity of the virus (40).
Apart from evaluating the roles of cleavage sites in transposition, systematic mutagenesis of the Ty1 cleavage sites also complements protein sequence data. Based on these results on Ty1 cleavage sites, the cleavage sites in Ty2, a related retroelement of S. cerevisiae, may be predicted. Although the Ty2 amino acid sequence diverges significantly from the Ty1 sequence in many places (36), sequences similar to the Ty1 cleavage sites can be readily identified (Table 2). The sequences of the six residues flanking the cleavage site differ by one to three residues. Most important, the hydrophobicity profiles of the matching cleavage sites in Ty1 and Ty2 are essentially the same, with P1 and P1′ amino acid residues being more hydrophilic than the surrounding P2 and P2′, consistent with the theory that hydrophobicity profiles and sequence accessibility in an unstructured region are the major determinants of cleavability (21, 30, 34).
TABLE 2.
Predicted sites of cleavage by Ty2 PR
| Site | Gag/PR | PR/IN | IN/RT |
|---|---|---|---|
| Ty1 sequence | KAH/NIA | TIN/NVH | LIA/AVK |
| Ty2 sequence | RAH/NVS | TIN/NVN | LIA/AIK |
PCS mutation phenotypes and trans complementation.
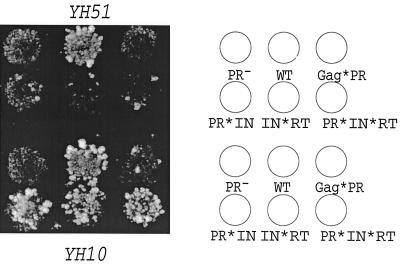
Transposition capabilities of the cleavage site mutants were evaluated in two congenic strains, YH10 and YH51 (Fig. 2 and 4). YH51 is an spt3 strain lacking a transcription factor required for expression of genomic Ty1 elements. Genomic Ty1 transposition is reduced 20-fold (37), and no Ty1-VLPs are made in spt3 strains unless a GAL-Ty1 plasmid is introduced (3). Unlike the native Ty1 promoter, the GAL promoter is SPT independent. Previous studies have shown that GAL-Ty1 elements with mutations in IN and RT are complemented at low levels by genomic Ty1 elements in SPT3+ strains (10). In contrast, PR active site mutants are not complemented in SPT3+ strains. These studies suggest that IN and RT mutants can be complemented in trans at a low level, whereas PR mutants cannot. We tested how our mutants behaved in these assays. For all cleavage site mutants, the transposition frequencies were at least 100 times lower than those of the wild-type Ty1-neo in the spt3 strain YH51. However, in the SPT3+ strain YH10, transposition was inhibited about 100-fold in cells expressing the Gag*PR mutant but only 3- to 4-fold in cells with PR*IN, IN*RT, or PR*IN*RT mutants. A simple explanation of these results is that genomic Ty1 elements can complement some but not all cleavage site mutations, presumably by providing correctly processed IN and or RT in trans. Alternatively, these mutations might produce proteins with dominant negative effects. Furthermore, as both the PR− and Gag*PR mutants fail to be complemented by the wild type, this result provides corroborating evidence that the primary transposition defect in the Gag*PR mutant is a protease defect.
FIG. 4.
Transposition assays. Yeast strain YH10 or YH51 cells were transformed to Ura+ with plasmids carrying wild-type (WT) and mutant Ty1 retroelements. Transformants were patched on SC-Ura glucose plates, then replica plated onto SC-Ura galactose plates, and incubated at 22°C for 48 h to induce transposition. The patches were then replica plated onto YPD nonselective medium to allow loss of the donor plasmid. Cells that lost the donor plasmid were selected by replica plating to SC–5-fluoro-orotic acid glucose medium and finally replica plated onto YPD medium containing 75 μg of G418 per ml to select for the cells whose genomes had acquired Ty1-neo. (Left) Growth on YPD medium containing G418; (right) diagram showing positions of mutant strains on the plate.
The PR/IN cleavage site functions normally in the place of the Gag/PR cleavage site.
To evaluate the potential significance of the primary sequence as opposed to the genomic position for the differential recognition and processing of the Ty1 PR cleavage sites, the six residues comprising the critical Gag/PR cleavage site were replaced by the equivalent residues of the PR/IN cleavage site, producing the PCS swap mutant (Fig. 2). Unlike the native Ty1 element, in which all three cleavage sites have different sequences, in the swap mutant the Gag/PR cleavage site is replaced by the PR/IN cleavage site. Ty1 protein processing was indistinguishable from the wild type in the swap mutant (Fig. 3C), as were the transposition frequencies (Fig. 2). Hence, at least the Gag/PR cleavage site can be substituted by another Ty1 cleavage site with little effect on processing or transposition. Most likely, the Gag/PR site is cleaved first not because of specific primary sequence of this site but because of its location at the N terminus of the protease.
In contrast, we also relocated the mature Gag/PR cleavage sites to positions 20 aa N terminally and 15 aa C terminally in constructs pM98 and pM100, respectively. In these constructs, no Gag processing was observed (Fig. 3), suggesting that position and hence three-dimensional structural context plays an important role in cleavage site selection, and that appropriate primary peptide sequence is necessary but not sufficient for cleavage in vivo.
Biochemical defects of the cleavage site mutants.
The biochemical basis for the mutant phenotypes of the PCS mutants has several potential explanations. It is possible that IN, when fused to PR or RT, does not translocate into the nucleus properly. The nuclear localization signal (NLS) has been mapped to the C-terminal part of Ty1 IN. The IN C terminus might need to be freed proteolytically from RT for efficient translocation to take place (20, 33). Alternatively, IN may be unable to integrate Ty1 DNA into genomic DNA in the nucleus as a result of its fusion to either PR or RT. Although it is possible that the RT is damaged in some way in the IN*RT mutant, Ty1 RT was previously shown to be active in in vitro homopolymer assays in the form of the Gag-Pol precursor found in the PR− mutant (39). However, more stringent tests of RT function are needed to ensure that RT does not have some more subtle polymerization defect.
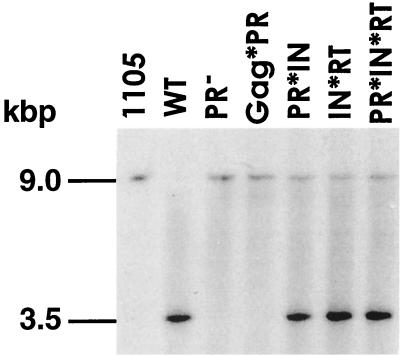
We investigated the ability of these mutants to make VLPs and also to synthesize Ty1 cDNA in vivo. The PCS mutants were capable of making VLPs in normal amounts, with readily detectable RT activity on exogenous primer templates. The PR− mutant and the Gag*PR mutant both failed to make Ty1 cDNA, whereas the PR*IN, IN*RT, and PR*IN*RT mutants all made amounts of Ty1 cDNA similar to wild-type levels (Fig. 5). These results support and extend earlier studies that showed that PR mutants fail to reverse transcribe endogenous Ty1 RNA effectively, presumably because they fail to access the endogenous primer or template but not because of an inactivity of the RT itself (39). In contrast, the PR*IN, IN*RT, and PR*IN*RT mutants appear to have a post-reverse transcription defect. Since integrase is affected by all three of these mutations, we expect that either transport of the preintegration complex to the nucleus, proper multimerization of IN, access to its preferred sites, or efficient concerted integration of the two ends of the Ty1 cDNA is blocked by these mutations.
FIG. 5.
Analysis of cDNA produced by the mutant elements. Total yeast nucleic acids prepared from galactose-induced cells were digested with EcoRI, RNase A treated, and electrophoretically separated on a 1% agarose gel. The positions of relevant molecular weight standards are indicated. The larger, 9.0-kbp band corresponds to the Ty1 donor plasmid; the smaller, 3.5-kbp band is derived from full-length Ty1-neo cDNA. Lane 1 shows donor plasmid (pJEF1105) alone. The blot was probed with a 32P-labeled neo cDNA probe. WT, wild type.
Evidence for a semiordered pathway of proteolytic cleavage in Ty1.
A polyprotein with more than one cleavage site may be processed into smaller products via a random process or via an ordered pathway in which the cleavage sites are cleaved sequentially. Ordered (or semiordered) processing had been well documented and is considered to be a common feature among retroviruses (11, 35). There is no simple universal pattern of ordered processing in retroviruses; rather, each virus has its own idiosyncratic pattern of cleavage. In general, however, cleavage of certain sites closer to the N terminus of the polyprotein precedes processing at more distal sites, as shown for avian sarcoma-leukemia virus (7, 13), murine leukemia virus (27), and HIV-1 (15, 18, 22). For instance, the N terminus of the retroviral CA is released before the C terminus (35). The order of Ty1 processing is consistent with this general retroviral trend, even though Ty1 biology is quite different.
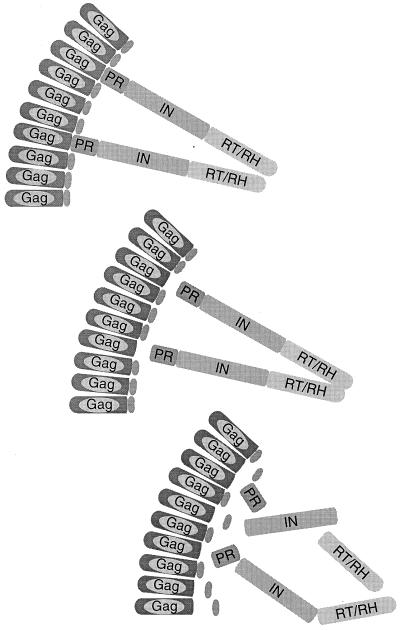
Cleavage sites of retrotransposons are not as well studied as their viral counterparts. The PR cleavage sites of Ty1, a copia-like element of yeast and of Ty3, a gypsy-like element, were previously determined by N- or C-terminal sequencing of mature proteins found in VLPs (6, 21, 30, 32). No cleavage sites have been mapped in other known retrotransposons. Systematic mutagenesis of Ty1 cleavage sites underscores the differences in proteolysis at these three sites and their unique roles in Ty1 processing and propagation. Moreover, the phenotypes of these mutations provide evidence for a semiordered processing pathway of Ty1 polypeptides. The Gag/PR cleavage site is cleaved first, and its cleavage is required for subsequent processing at the PR/IN and IN/RT sites (Fig. 6). This is consistent with results of pulse-chase experiments on Ty1 processing done in vivo (17). Our data do not indicate an obligatory order of processing at the other two sites; rather, it is clear that cleavage of PR/IN is not required for the cleavage of IN/RT, and vice versa. Elucidation of the processing sites in other retrotransposons and their mutagenesis may provide valuable insights into the mechanisms and regulation of proteolytic processing and retrotransposition.
FIG. 6.
Model for ordered processing pathway in Ty1 VLP assembly. Sections of a Ty1 VLP are shown. Shaded boxes, Ty1 proteins. Top, Ty1 proteins before cleavage; middle, the first step of processing, autocatalytic cleavage of the Gag/PR site; bottom, processing of PR/IN and IN/RT sites.
ACKNOWLEDGMENTS
We thank G. Sharon and David Garfinkel for sharing unpublished observations on Ty1 PR cleavage site mutants, and we thank Jeff Smith for helpful suggestions.
This work was supported in part by NIH grant GM36481 to J.D.B. and Medical Scientist training grant GM-07309 to J.F.L.
REFERENCES
- 1.Boeke J D, Garfinkel D J, Styles C A, Fink G R. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 2.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Varmus H, Hughes S, Coffin J, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 3.Boeke J D, Styles C A, Fink G R. Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Mol Cell Biol. 1986;6:3575–3581. doi: 10.1128/mcb.6.11.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke J D, Xu H, Fink G R. A general method for the chromosomal amplification of genes in yeast. Science. 1988;239:280–282. doi: 10.1126/science.2827308. [DOI] [PubMed] [Google Scholar]
- 5.Braiterman L T, Monokian G M, Eichinger D J, Merbs S L, Gabriel A, Boeke J D. In-frame linker insertion mutagenesis of yeast transposon Ty1: phenotypic analysis. Gene. 1994;139:19–26. doi: 10.1016/0378-1119(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 6.Braiterman T L. Ph.D. thesis. Baltimore, Md: The Johns Hopkins University; 1993. [Google Scholar]
- 7.Burstein H, Bizub D, Kotler M, Schatz G, Vogt V M, Skalka A M. Processing of an avian retroviral Gag polyprotein precursors is blocked by a mutation at the NC-PR cleavage site. J Virol. 1992;66:1781–1785. doi: 10.1128/jvi.66.3.1781-1785.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron J R, Loh E Y, Davis R W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979;16:739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- 9.Clare J, Belcourt M, Farabaugh P J. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc Natl Acad Sci USA. 1988;85:6816–6820. doi: 10.1073/pnas.85.18.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curcio M J, Garfinkel D J. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol Cell Biol. 1992;12:2813–2825. doi: 10.1128/mcb.12.6.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson C, Eisenman R, Fan H, Hunter E, Teich N. Protein biosynthesis and assembly. In: Weiss R, editor. Molecular biology of tumor viruses: RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 513–648. [Google Scholar]
- 12.Eichinger D J, Boeke J D. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988;54:955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 13.Eisenman R, Mason W S, Linial M. Synthesis and processing of polymerase proteins of wild-type and mutant avian retroviruses. J Virol. 1980;36:62–78. doi: 10.1128/jvi.36.1.62-78.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elder R T, St. John T P, Stinchcomb D T, Davis R W. Studies on the transposable element Ty1 of yeast I. RNA homologous to Ty1. Cold Spring Harbor Symp Quant Biol. 1981;45:581–584. doi: 10.1101/sqb.1981.045.01.075. [DOI] [PubMed] [Google Scholar]
- 15.Erickson-Viitenan S, Manfredi J, Viitanen P, Tribe D E, Tritch R, Hutchison III C A, Loeb D D, Swanstrom R. Cleavage of HIV-1 Gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res Hum Retroviruses. 1989;5:577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 16.Garfinkel D J, Boeke J D, Fink G R. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985;42:507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 17.Garfinkel D J, Hedge A-M, Youngren S D, Copeland T D. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991;65:4573–4581. doi: 10.1128/jvi.65.9.4573-4581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gowda S D, Stein B S, Engleman E G. Identification of protein intermediates in the processing of the p55 HIV-1 Gag precursor in cells infected with recombinant vaccinia virus. J Biol Chem. 1989;264:8459–8462. [PubMed] [Google Scholar]
- 19.Kawakami K, Pande S, Faiola B, Moore D P, Boeke J D, Farabaugh P J, Strathern J N, Nakamura Y, Garfinkel D J. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics. 1993;135:309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenna M A, Brachmann C B, Devine S E, Boeke J D. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol. 1998;18:1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchner J, Sandmeyer S. Proteolytic processing of Ty3 proteins is required for transposition. J Virol. 1993;67:19–28. doi: 10.1128/jvi.67.1.19-28.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krausslich H G, Schneider H, Zybarth G, Carter C A, Wimmer E. Processing of in vitro synthesized Gag precursor proteins of human immunodeficiency virus (HIV) type 1 by HIV proteinase generated in Escherichia coli. J Virol. 1988;62:4393–4397. doi: 10.1128/jvi.62.11.4393-4397.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leis J, Baltimore D, Bishop J M, Coffin J, Fleissner E, Goff S P, Oroszlan S, Robinson H, Skalka A M, Temin H M, Vogt V. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeb D D, Huchinson III C A, Edgell M H, Farmerie W G, Swanstrom R. Mutational analysis of human immunodeficiency virus type 1 protease suggests functional homology with aspartic proteinases. J Virol. 1989;63:111–121. doi: 10.1128/jvi.63.1.111-121.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louis J M, Nashed N T, Parris K D, Kimmel A R, Jerina D M. Kinetics and mechanism of autoprocessing of human immunodeficiency virus type 1 protease rom an analog of the Gag-Pol polyprotein. Proc Natl Acad Sci USA. 1994;91:7970–7974. doi: 10.1073/pnas.91.17.7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maxwell S, Arlinghaus R B. In vitro proteolytic cleavage of Gazdar murine sarcoma virus p65gag. J Virol. 1981;39:963–967. doi: 10.1128/jvi.39.3.963-967.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellor J, Fulton A M, Dobson M J, Roberts N A, Wilson W, Kingsman A J, Kingsman S M. The Ty transposon of Saccharomyces cerevisiae determines the synthesis of at least three proteins. Nucleic Acids Res. 1985;13:6249–6263. doi: 10.1093/nar/13.17.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellor J, Malim M H, Gull K, Tuite M F, McCready S, Dibbayawan T, Kingsman S M, Kingsman A J. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature. 1985;318:583–586. doi: 10.1038/318583a0. [DOI] [PubMed] [Google Scholar]
- 30.Merkulov G V, Swiderek K M, B. B C, Boeke J D. A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J Virol. 1996;70:5548–5556. doi: 10.1128/jvi.70.8.5548-5556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monokian G M, Braiterman L T, Boeke J D. In-frame linker insertion mutagenesis of yeast transposon Ty1: mutations, transposition and dominance. Gene. 1994;139:9–18. doi: 10.1016/0378-1119(94)90517-7. [DOI] [PubMed] [Google Scholar]
- 32.Moore S P, Garfinkel D J. Expression and partial purification of enzymatically active recombinant Ty1 integrase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:1843–1847. doi: 10.1073/pnas.91.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore S P, Rinckel L A, Garfinkel D J. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol. 1998;18:1105–1114. doi: 10.1128/mcb.18.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettit S C, Simsic J, Loeb D D, Everitt L, Hutchison C A, Swanstrom R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J Biol Chem. 1991;266:14539–14547. [PubMed] [Google Scholar]
- 35.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 36.Warmington J R, Waring R B, Newlon C S, Indge K J, Oliver S G. Nucleotide sequence characterization of Ty 1-17, a class II transposon from yeast. Nucleic Acids Res. 1985;13:6679–6693. doi: 10.1093/nar/13.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winston F, Durbin K J, Fink G R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984;39:675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- 38.Xu H, Boeke J D. Inhibition of Ty1 transposition by mating pheromones in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2736–2743. doi: 10.1128/mcb.11.5.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Youngren S D, Boeke J D, Sanders N J, Garfinkel D J. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol Cell Biol. 1988;8:1421–1431. doi: 10.1128/mcb.8.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zybarth G, Krausslich H G, Partin K, Carter C. Proteolytic activity of novel human immunodeficiency virus type 1 proteinase proteins from a precursor with a blocking mutation at the N terminus of the PR domain. J Virol. 1994;68:240–250. doi: 10.1128/jvi.68.1.240-250.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]