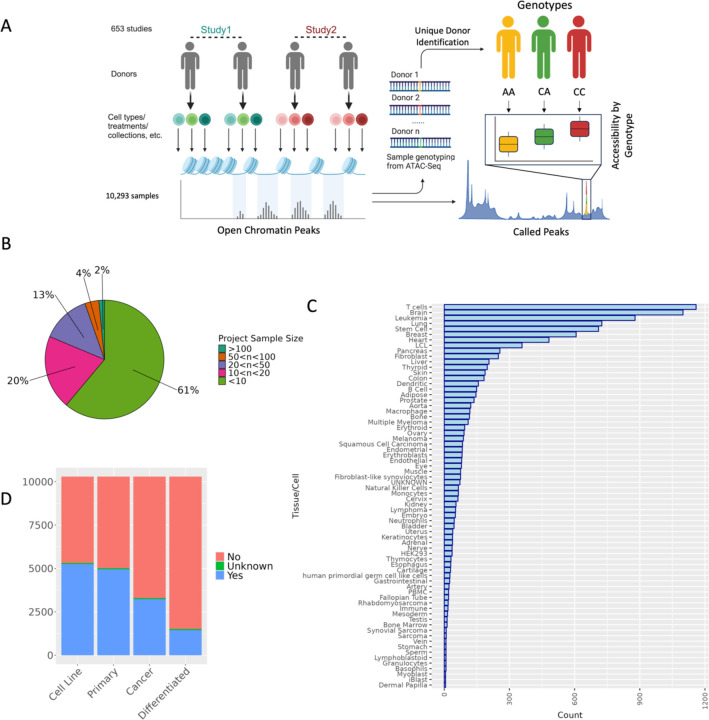
Figure 1. Study overview and characteristics of specimens utilized in this study.
(A) Overview of study design to jointly call genotype and caQTLs across studies. Human ATAC-seq datasets were obtained from GEO. After variant-calling (Methods), we identified the unique donors in the dataset (Methods) for use in caQTL mapping. (B) The distribution of the number of samples collected across all n=653 studies. (C) Frequency of the Cell/Tissue types present in samples collected across studies based on manual metadata curation (D) Frequencies of cancer, non-cancer, primary tissues, and cell-line samples included in our study based on our metadata review. For each category, samples were assigned a “Yes” if they belonged to that category (e.g. cell line samples for ‘Cell Line’ category), a “No” if they did not belong (e.g. primary tissue samples for ‘Cell Line’ category), or an “Unknown” if it was not clear from the metadata.