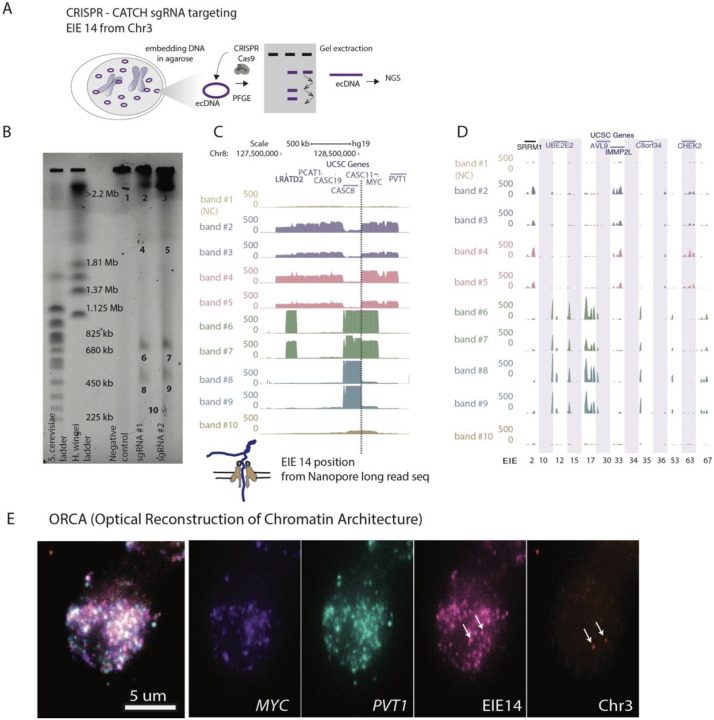
Figure 2: CRISPR-CATCH Elucidates ecDNA Composition and EIE Insertions.
A. Schematic diagram illustrating the CRISPR-CATCH experiment designed to isolate and characterize ecDNA components. The process involves the use of guide RNA targeting the EIE 14 from Chromosome 3. DNA is embedded in agarose, followed by pulse-field gel electrophoresis (PFGE), allowing for the band extraction and subsequent next-generation sequencing (NGS) of ecDNA fragments.
B. PFGE gel image displays the separation of DNA fragment, lines from left ladder, ladder, empty lane, Negative control, sgRNA #1, sgRNA #2 and band numbers for NGS. The EIE 14 targeted by the guide RNAs leads to cutting of the ecDNA's chromosome 8 sequences to form multiple discrete bands. sgRNA #1 ATATAGGACAGTATCAAGTA; sgRNA #2 TATATTATTAGTCTGCTGAA; Full EIE 14 sequences from long-read sequencing is in Supplementary Table T6.
C. Visualization of the sequencing results confirms the presence of EIE 14, originally annotated on Chromosome 3, within the ecDNA, between the CASC8 and CASC11 genes, approximately 200 kilobases upstream from MYC. The dotted line indicates the position of this insertion.
D. Additional EIEs identified in the initial Hi-C screen, captured, and sequenced in the CRISPR-CATCH, each EIE is one one vertical shaded box with coordinates.
E. ORCA (Optical Reconstruction of Chromatin Architecture) visualization of the COLO320DM cell nucleus. The images show the spatial arrangement of the MYC oncogene, EIE 14 and the PVT1 locus, labeled in different colors. The scale bar represents 5 micrometers. Chr3 probe maps to the breakpoints of the EIE 14 origin inside CD96 intron.