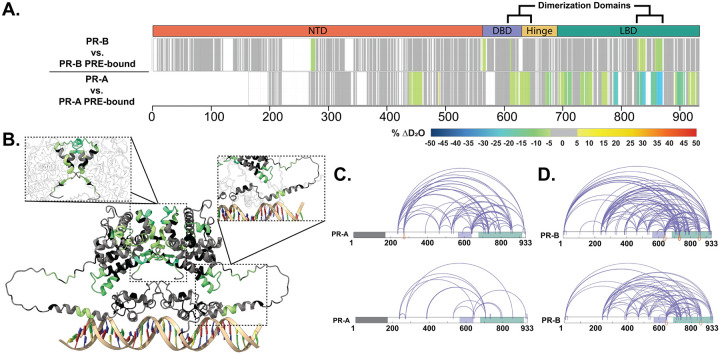
Figure 2. Structural proteomics reveals isoform differences upon response element binding.
A. Consolidated HDX-MS data, run in triplicate, showing the differential analysis between unbound PR vs. PRE-bound where the top is PR-B, and the bottom is PR-A. Domains are labeled as the following: N-terminal Domain (NTD), DNA-binding domain (DBD), Hinge region (Hinge), and Ligand binding domain (LBD). B. Trimmed AlphaFold 3.0 model (residues 375–769) of PR-A homodimer with unbound PR-A vs. PR-A:PRE HDX overlays. Highlighted regions are the PR dimerization domain (top) and the DBD C-terminal extension (bottom). Cooler colors indicate stronger HDX protection. C. XlinkX images of differential PR-A ± PRE experiments, where crosslinks from the unbound (top) and PRE-bound (bottom) states are shown. Crosslinks mapped onto PR-B numbering with the gray region representing the 164 amino acids not expressed in PR-A. Results representative of triplicate experiments, with validation in Skyline. D. XlinkX view of differential PR-B ± PRE experiments, where crosslinks from unbound (top) and PRE-bound (bottom) states are shown. Domains – Gray: Not expressed; Purple: DBD; Green: LBD.