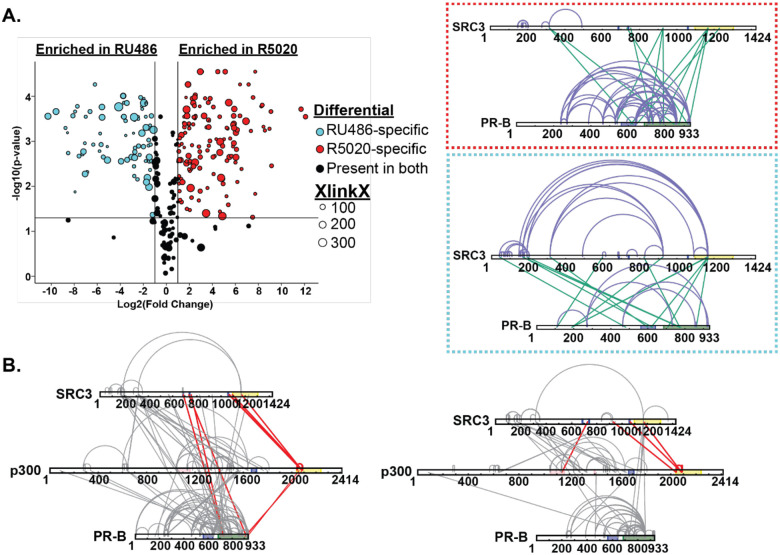
Figure 7. RU486 antagonism rearranges PR-SRC3-p300 interactions.
Top. Plotted differential crosslinks in PR:SRC3 experiments, comparing R5020-specific (agonist, red) and RU-486-specific (antagonist, blue) crosslinks. Crosslinks represented by circles with corresponding XlinkX scores as point size. Top. Red. XlinkX view of R5020-specific crosslinks in differential PR:SRC3 experiments. Top. Blue. XlinkX view of RU486-specific crosslinks in differential PR:SRC3 experiments. B. All validated R5020-bound (left) and RU486-bound (right) crosslinks for differential PR-B:SRC3:p300 ± PRE experiments. The x-axis represents the Log2 transformed fold change values from Skyline, while the y-axis represents the −log10 transformation of the Skyline p-value output. The lines are indicative of a Log2 fold change of 1 (two-fold increase) and −log10 p-value of 1.3, corresponding to p<0.05. Crosslinks highlighted in red show notable differences in PR:SRC3:p300 interactions between PR ligands. Defined domains are as follows: PR - DBD (purple) and LBD (green); SRC3 – NR-boxes (purple) and histone acetyltransferase domain (yellow); p300 - bromodomain (pink), zinc finger domain (green), and NCOA2-interaction domain (yellow).