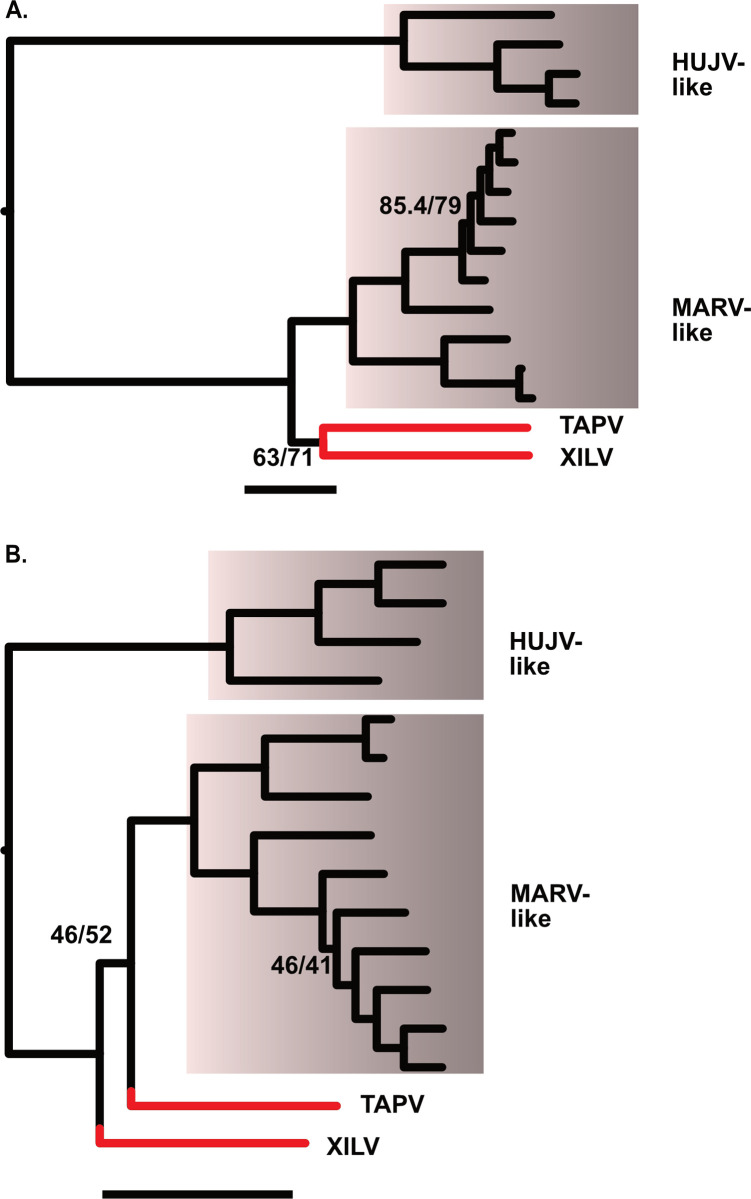
Fig 1. Phylogenies showing relationships of four deep lineages/clades of known filoviruses.
The x-axis for each graph is proportional in length to genetic distances (the scale bar is 1 substitution/site). Acronyms are Huángjiāo virus (HUJV), Marburg virus (MARV), Tapajós virus (TAPV), and Xīlǎng virus (XILV). Note that TAPV and XILV form a long branch pair with the amino acid data. Numbers represent branches with the lowest approximate likelihood ratio tests and bootstrap values. The remaining branches had support greater than 96. A. ML tree based on the L Protein amino acid sequence alignment B. ML tree based on partitioned codon model (nucleotides) for the same alignment as in A.