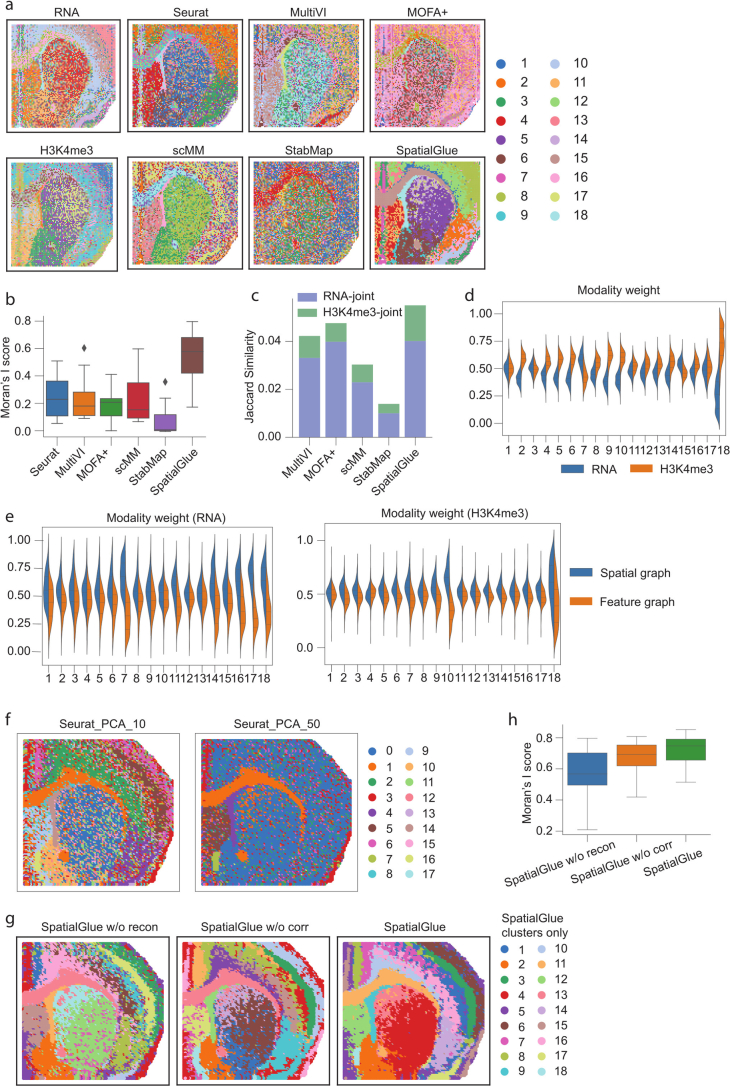
Extended Data Fig. 5. Results of the mouse brain P22 sample acquired with RNA-seq and CUT&Tag-seq (H3K4me3).
(a) Spatial plots of data modalities with unimodal clustering (left), and clustering results (right) from single-cell and spatial multi-omics integration methods, Seurat, MultiVI, MOFA+, scMM, StabMap, and SpatialGlue. (b) Comparison of Moran’s I score. In the box plot, the center line denotes the median, box limits denote the upper and lower quartiles, and whiskers denote the 1.5× interquartile range. n = 18 clusters. (c) Comparison of Jaccard Similarity scores. (d) Between-modality weights explaining the importance of each modality to each cluster. (e) Within-modality weights explaining the contributions of the spatial and feature graphs to each cluster for each modality. (f) Comparison of spatial clustering using Seurat with 10 and 50 PC dimensions in the mouse brain P22 spatial-ATAC-RNA-seq sample. (g) Comparison of SpatialGlue and its variants, that is, SpatialGlue without reconstruction loss (‘SpatialGlue w/o recon’) and SpatialGlue without correspondence loss (‘SpatialGlue w/o corr’), in the mouse brain P22 spatial-ATAC-RNA-seq sample. (h) Comparison of Moran’s I score of SpatialGlue and its two variants. In the box plot, the center line denotes the median, box limits denote the upper and lower quartiles, and whiskers denote the 1.5× interquartile range. n = 18 clusters.