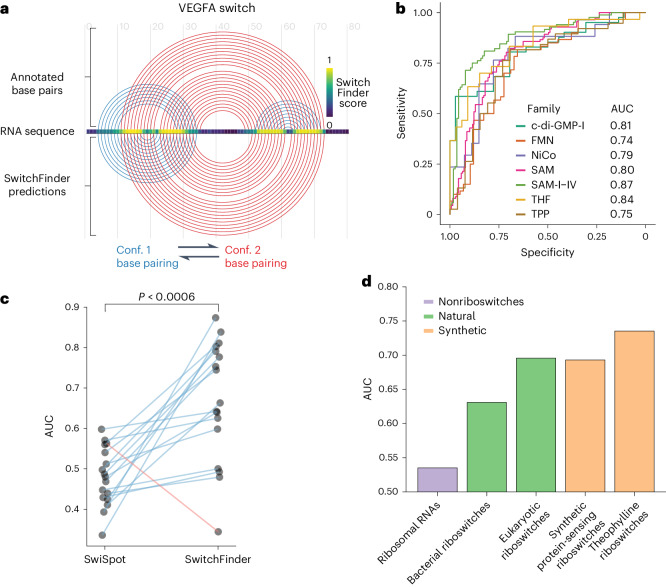
Fig. 1. SwitchFinder identifies candidate RNA switches in the human genome.
a, Example of SwitchFinder locating the RNA switch in the VEGFA mRNA sequence. b, Receiver operating characteristic (ROC) curves of SwitchFinder predictions of RNA switches from the common Rfam families. SwitchFinder was applied to a mix of real sequences and their shuffled counterparts (with preserved dinucleotide content). ROC curves measure its ability to correctly select the real sequences. AUC, area under the ROC curve; riboswitch families, c-di-GMP-I (Cyclic di-GMP); FMN, flavin mononucleotide; NiCo, nickel or cobalt ions; SAM, S-adenosyl-l-methionine; THF, tetrahydrofolate; TPP, thiamine pyrophosphate. c, AUCs of RNA switch predictions across the Rfam families for two models: SwitchFinder and SwiSpot10. Each dot represents one Rfam family. The lines show the change in accuracy between the two models. The families that have higher AUCs for SwitchFinder are shown with blue lines; the ones that have higher AUCs for SwiSpot are shown in red. P value calculated with the paired two-sided t-test (P = 0.00056). d, AUCs of RNA switch predictions across various groups of natural and synthetic riboswitches, calculated as in b.