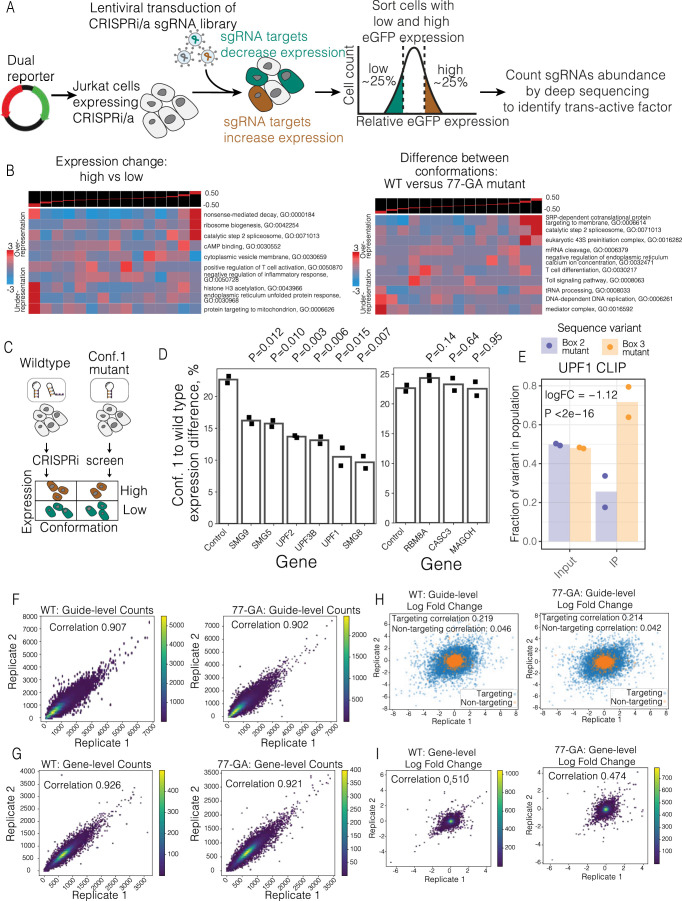
Extended Data Fig. 7. CRISPRi screen highlights the pathways acting downstream of the RORC RNA switch.
a Overview of the flow cytometry-based CRISPRi screen workflow. b Gene set enrichment analysis of the data depicted in Fig. 6a (left) and Fig. 6b (right). The genes were distributed into equally populated bins based on their comparative abundance between high expression and low expression quartiles (left), or based on their comparative phenotype in the CRISPRi screens performed in WT or 77-GA mutant backgrounds (right). Then the enrichment of a given gene set was calculated in each bin using iPAGE, a mutual information-based algorithm (Goodarzi et al. 2009). c Experiment design table. d The effect of knockdown of SURF and EJC complex member proteins on the expression change upon the conformation equilibrium shift. The individual genes were knocked down using the CRISPRi system in both WT and 77-GA mutant cell lines, then the change of reporter gene expression was measured by flow cytometry (N replicates = 2). The bar plots demonstrate the expression ratios of WT to 77-GA mutation cell lines. e The bar plots demonstrate the fractions of reads carrying the Box 2 (77-GA) mutant sequence or Box 3 (116-CCCTAAG) mutant sequence in UPF1 cross-linking and immunoprecipitation (CLIP) library. Box 2 mutant favors conformation 1, Box 3 mutant favors conformation 2. Left: input RNA libraries, extracted from the Box 3 and Box 2 mutant-expressing Jurkat cells, mixed at 1:1 ratio. Right: libraries after anti-UPF1 immunoprecipitation. P value was calculated using Translation Efficiency Ratio test as in (Navickas et al. 58). N replicates = 2. f Density plots showing the correlation of sgRNA counts between the replicates of the CRISPRi screens performed in the WT (left) and 77-GA mutant (right) backgrounds. g Density plots showing the correlation of gene counts between the replicates of the CRISPRi screens performed in the WT (left) and 77-GA mutant (right) backgrounds. The counts of all the sgRNAs targeting a given gene are pooled and reported as a single number (N = 5 sgRNAs per gene). h Scatter plots showing the correlation of sgRNA phenotypes between the replicates of the CRISPRi screens performed in the WT (left) and 77-GA mutant (right) backgrounds. Logarithmic fold changes between the sgRNA abundance ‘high’ and ‘low’ expression bins are shown on both axes. Nontargeting sgRNAs are shown in orange; all the other sgRNAs are shown in blue. The correlation values are reported separately for nontargeting and targeting sgRNAs. i Density plots showing the correlation of gene phenotypes between the replicates of the CRISPRi screens performed in the WT (left) and 77-GA mutant (right) backgrounds. Logarithmic fold changes between the abundance of sgRNAs targeting a given gene in ‘high’ and ‘low’ expression bins are shown on both axes.