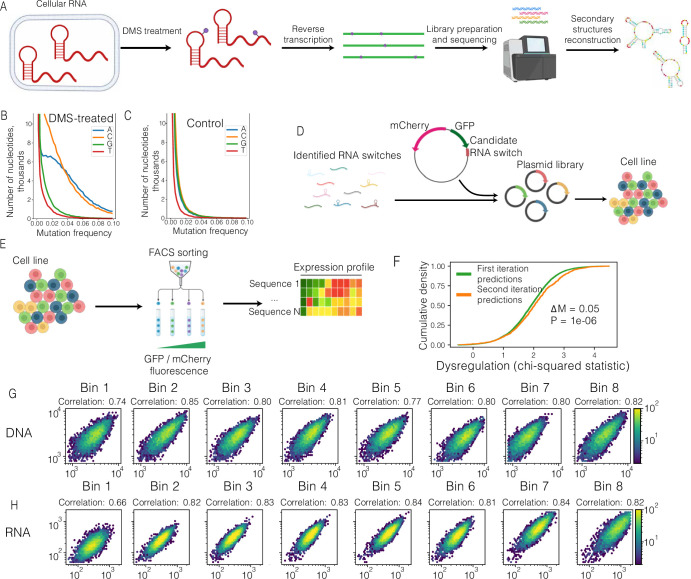
Extended Data Fig. 2. Overview of high-throughput screening approaches for improved RNA switch predictions.
a Overview of DMS-MaPseq workflow. Mammalian cells are treated with DMS. DMS-modified nucleotides cause mutations when cDNA is synthesized from RNA templates. The cDNA libraries are sequenced, the DMS-caused mutations are counted, providing the Watson-Crick face accessibility estimates for each A- or C- nucleotide. b Cumulative mutation frequency in DMS-treated candidate RNA switches, separated by nucleotide. c Cumulative mutation frequency in nontreated candidate RNA switches, separated by nucleotide. d Overview of the library generation workflow for Massively Parallel Reporter Assay (MPRA). Sequences of candidate RNA switches are synthesized as DNA oligonucleotides and cloned into a reporter vector into 3ʹUTR region of a eGFP cDNA. The plasmid library is packaged into lentiviral particles, and used for infecting mammalian cells. The infection is performed at low MOI (infection rate) to ensure that most cells get only a single plasmid copy. e Overview of the MPRA workflow. A population of mammalian cells is separated into bins based on GFP/mCherry fluorescence ratio. In the schematic, cells are colored according to the sequence they carry in the 3ʹUTR of the GFP reporter. f Cumulative density plot of dysregulation values, comparing the candidate RNA switches predicted in first and second (DMS-MaPseq informed) iterations of SwitchFinder. Dysregulation values are estimated using chi-square test for every individual candidate RNA switch across 8 expression bins. Median difference (∆M) and P value (calculated using Mann-Whitney U-test) are shown. g Correlations of read counts of gDNA libraries between the biological replicates of massively parallel mutagenesis analysis. h Correlations of read counts of RNA libraries between the biological replicates of massively parallel mutagenesis analysis.