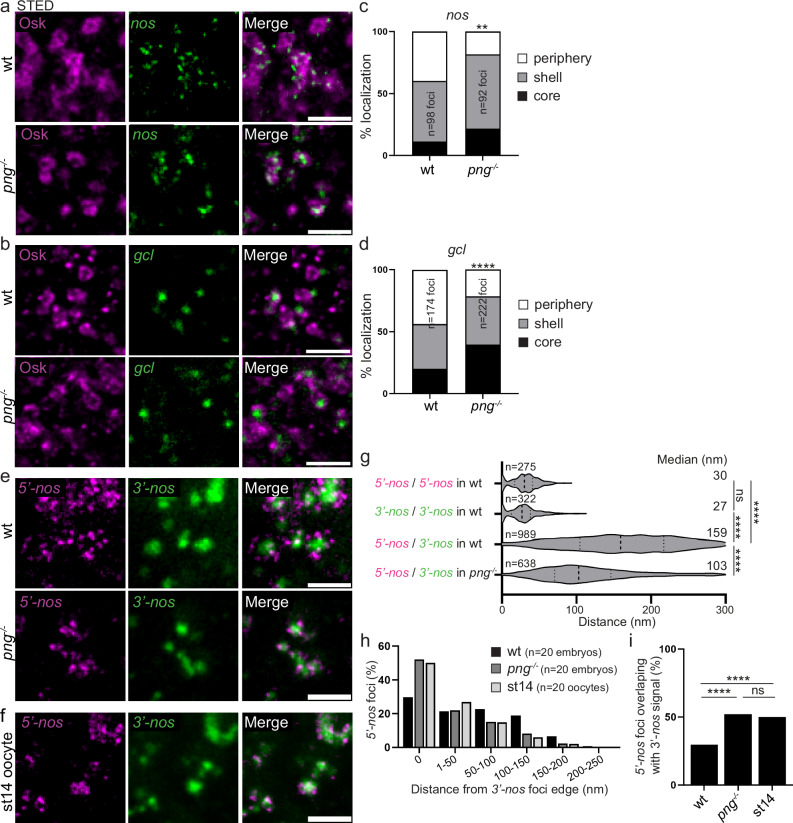
Fig. 6. mRNA localization and compaction within germ granules depend on their translation.
a, b STED imaging of immuno-smFISH of wild-type (top) and png1058 (bottom) embryos with anti-Osk antibody (magenta) as a marker of germ granule and smFISH probes (green) against nos (a) and gcl (b) mRNAs. c, d Percentage of localization of nos (c) and gcl (d) mRNA foci in wild-type and png1058 mutant embryos, in the core (black), in and at the surface of the shell (grey), and at the immediate periphery (white) of germ granules from images as in (a, b). **p < 0.01, ****p < 0.0001 using the χ2 test. p = 0.0031 in (c) and 1.75 × 10−22 in (d). e STED images of smFISH against nos 5′end (5′-nos, magenta) and 3′end (3′-nos, green) in wild-type and png1058 mutant embryos. f STED images of smFISH against nos 5′end (5′-nos, magenta) and 3′end (3′-nos, green) in wild-type stage 14 oocytes. g Violin plots showing distance distribution of colocalizing foci (5′-nos to 5′-nos and 3′-nos to 3′-nos) and 5′end to 3′end distances for nos mRNAs at germ granules in wild-type and png1058 mutant embryos, from STED images as in (e). Dashed lines inside the violin plots show first quartile, median, and third quartile. Median distances are indicated on the right. ns: non-significant, ****p < 0.0001 using unpaired two-tailed Student’s t-test. p = 0.2 between 5′-nos/5′-nos and 3′-nos/3′-nos, p = 3.5 × 10−191 between 5′-nos/5′-nos and 5′-nos/3′-nos in wt, p = 1.3 × 10−218 between 3′-nos/3′-nos and 5′-nos/3′-nos in wt and p = 4.5 × 10−58 between 5′-nos/3′-nos in wt and png−/−. The number of measured distances is indicated (n). h Measurement of the distance between nos 5′end foci and the edge of nos 3′end foci in wild-type embryos, png1058 embryos, and wild-type stage 14 oocytes (st14), from images as in (e, f). The histogram shows the percentage of nos 5′end foci in each distance class. i Percentage of nos 5′end foci overlaps with nos 3′end foci in wild-type embryos, png1058 embryos and wild-type stage 14 oocytes (st14). ns: non-significant, ****p < 0.0001 using the χ2 test. p = 2.64 × 10−129 between wt and png−/−, p = 1.79 × 10−167 between wt embryos and stage 14 oocytes, and p = 0.15 between png−/− embryos and stage 14 oocytes. Scale bars: 1 µm. Source data are provided as a Source Data file.