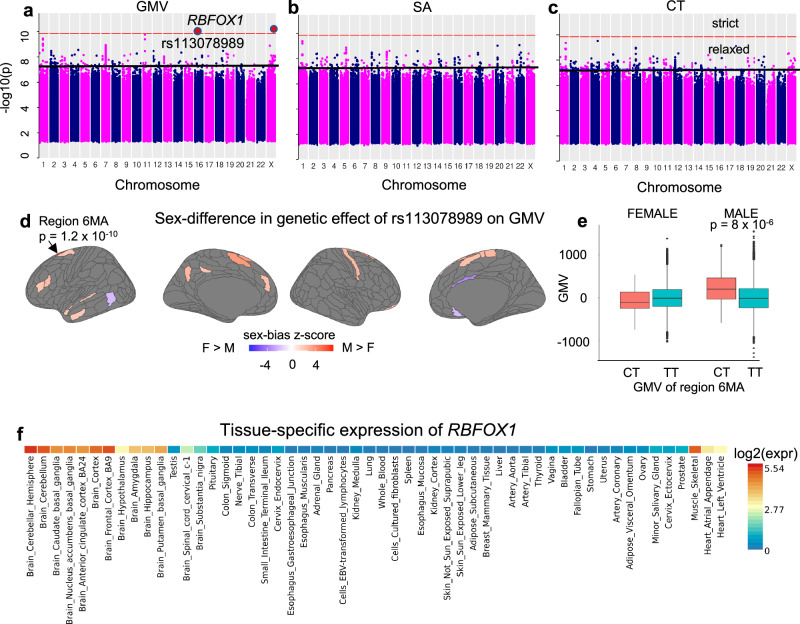
Fig. 4. Genome-wide tests of sex-difference in SNP effects in regional gray matter volume (GMV), surface area (SA), and cortical thickness (CT).
a–c Minimum p-value across all 360 cortical regions for each SNP for each phenotype category: GMV (a), SA (b), and CT (c). The red and the black horizontal lines correspond to the “strict” and the “relaxed” significance thresholds (pstrict = 1.4e-10, which accounts for multiple-testing and prelaxed = 5e-8, respectively). Red circles indicate SNPs above the “strict” threshold: chr16:rs113078989 (p = 1.2e-10) and chrX:rs747862348 (p = 9.5e-11). rs113078989 mapped to protein-coding gene RBFOX1. The sample sizes for the relevant sex-stratified genome-wide association analyses may be found in Supplementary Data 1. d Sex-difference in the effect of rs113078989 across the cortex on cortical GMV shown as z-scores (“Methods”) with red indicating higher magnitude in male participants; all regions with sex-difference p-value > 0.05 are shown in gray. e Boxplot showing GMV values for each sex (N = 13968 male, N = 15613 female) for the CT and TT genotypes of rs113078989 in region 6MA. The p-value for the difference in GMV between the two genotypes in the male group was calculated using a two-sided t test (N = 13968, t = − 4.7, df = 81.7, p = 8.2e-6, mean difference = − 214.17, 95% CI = [− 303.67, − 124.68]). In the boxplots, the center shows the median, and the lower and the upper hinges correspond to the first and third quartiles. The lower whisker extends from the hinge to the smallest value at most 1.5 * IQR of the hinge, where IQR is the distance between the first and the third quartile. The upper whisker extends from the hinge to the largest value no further than 1.5 * IQR from the hinge. f GTEx tissue expression heatmap plot for RBFOX1 indicating significant brain expression (from FUMA). The unit shown is log2 of the expression per label. Red indicates high expression. Source data are provided as Source Data files.