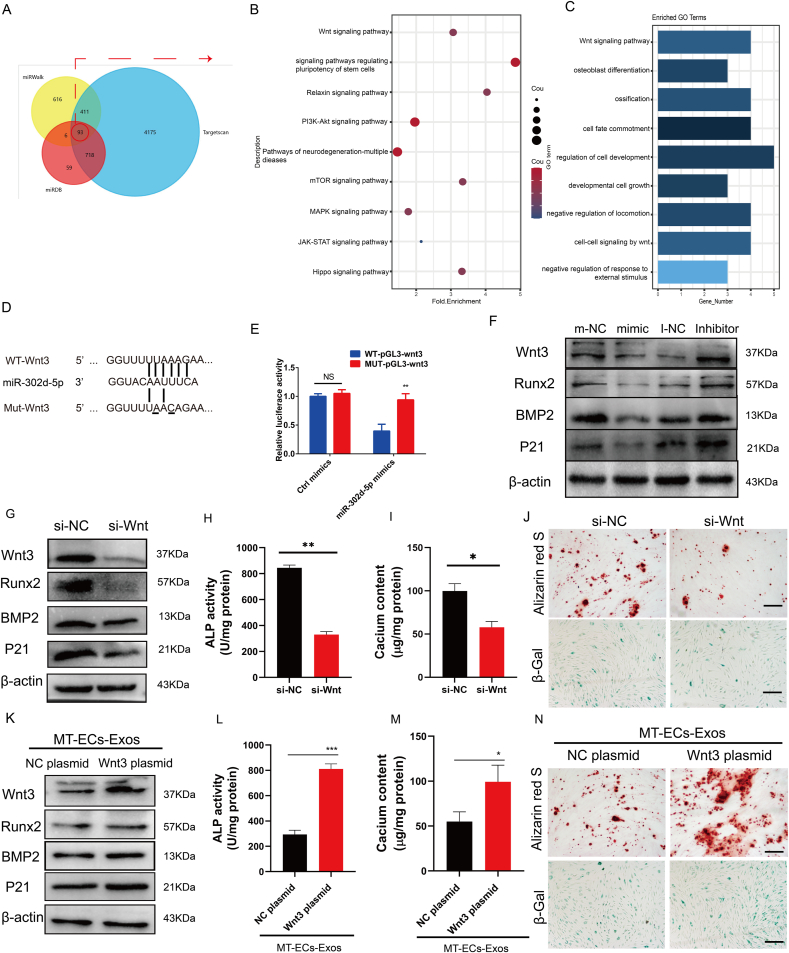
Fig. 5.
miR-302d-5p inhibits VSMCs calcification and senescence by targeting Wnt3 (A) The Venn diagram of target genes of miR-302d-5p predicted by TargetScan, miRwalk and miRDB. (B) Gene Ontology analysis of overlapping target genes of miR-302d-5p. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of overlapping target genes of miR-302d-5p. (D) Schematic representation of miR-302d-5p putative target sites in Wnt3 3′-UTR and alignment of miR-302d-5p with WT and MUT Wnt3 3′-UTR showing pairing. (E) Luciferase activity was performed using luciferase constructs carrying a wild type or mutant Wnt3 3′-UTR co-transfected into HEK293T cells with miR-302d-5p mimics compared with empty vector control. (F)The expression level of Wnt3, Runx2, BMP2, P21 in VSMCs treated with miR-302d-5p mimic or inhibitor. (G–J) VSMCs were incubated with Wnt3 silencing by siRNA interference. Then, the expression level of Wnt3, Runx2, P16, P21(G), ALP activity (H), calcium content (I) and mineralized nodules (J, upper panel, scale bar = 200 μm) as well as senescence cells (J, lower panel, scale bar = 200 μm) was measured in VSMCs. (K–N) VSMCs were transfected with Wnt3 overexpression plasmid before MT-ECs-Exos incubation. Then, the expression level of Wnt3, Runx2, P16, P21(K), ALP activity (L), calcium content (M) and mineralized nodules (N, upper panel) as well as senescence cells (N, lower panel) was measured in VSMCs (scale bar = 200 μm). Results are represented by mean ± SEM for each group. *p < 0.05. **p < 0.01, ***p < 0.001.