Abstract
Zinc is an essential micronutrient that cells must obtain from the environment in order to develop their normal growth. Previous work performed at our laboratory showed that the synthesis of immunodominant antigens from Aspergillus spp., including A. fumigatus, was up-regulated by a low environmental concentration of zinc. These results suggested that a tightly regulated system for the fungus to grow under zinc-limiting conditions must underlie the ability of A. fumigatus to acquire zinc in such environments. In this work, we show that zrfA and zrfB are two of the genes that encode membrane zinc transporters from A. fumigatus in this system. Expression of these genes is differentially down-regulated by increasing concentrations of zinc in the medium. Thus, the transcription of zrfB is turned off at a concentration 50-fold higher than that for zrfA transcription. In addition, phenotypic analyses of single zrfAΔ and zrfBΔ mutants and a double zrfAzrfBΔ mutant revealed that the deletion of zrfB causes a greater defect in growth than the single deletion of zrfA. Deletion of both genes has a dramatic effect on growth under acid, zinc-limiting conditions. Interestingly, in neutral or slightly alkaline zinc-depleted medium, the transcriptional expression of both genes is down-regulated to such an extent that even in the absence of a supplement of zinc, the expression of zrfA and zrfB is strongly reduced. This fact correlates with the growth observed in alkaline medium, in which even a zrfAzrfBΔ double mutant was able to grow in a similar way to the wild-type under extremely zinc-limiting conditions. In sum, the zinc transport proteins encoded by zrfA and zrfB are members of a zinc uptake system of A. fumigatus that operates mainly under acid, zinc-limiting conditions.
Zinc is the second most abundant transition metal in cells after iron. Although it is found in cells in trace amounts, this element is an essential component of many enzymes. More than 300 different enzymes, including representatives of all six major functional enzyme classes, use zinc as a cofactor for their catalytic activity or/and structural stability by mediating the correct folding of specific domains (1). Thus, zinc is considered to be an essential micronutrient for all organisms, although excess amounts of zinc may prove poisonous for cells. As a consequence, a tight regulation of its availability is a critical issue in the normal growth of all organisms. However, as a highly charged metal ion, zinc cannot passively enter the cell through the hydrophobic lipid bilayer of the plasma membrane but instead requires specific membrane transporter proteins for its incorporation into cells (10).
Aspergillus fumigatus is a saprophytic fungus that inhabits soils, where it grows on organic debris and releases hundreds of conidia into the atmosphere. Conidia inhaled by immunocompetent individuals are efficiently eliminated by nonspecific immune cells, which constitute the first defense barrier (19). However, in the lung tissue of immunocompromised patients, extensive growth of A. fumigatus may result in invasive aspergillosis, the most fatal variety of aspergillosis (22). Wherever A. fumigatus grows (in soil or in living tissue), its survival depends on its ability to obtain all the nutrients, including essential micronutrients from the environment such as iron, copper, or zinc, needed to support its growth. Since A. fumigatus, like most microorganisms, generally lives in environments that are nutrient limiting due to low initial availability or depletion caused by the growth of any other microorganism, this fungal pathogen has developed different mechanisms to obtain metal nutrients from its medium. In this context, much work has been done regarding the mechanism of iron uptake by pathogenic microorganisms (18). Aspergillus spp., including pathogenic strains of A. fumigatus, excrete different kinds of siderophores to mobilize extracellular iron with high affinity through siderophore transporters (23, 26). The acquisition of zinc has also been recognized as one of the key steps in the process of infection by any pathogen, since this metal is strongly sequestered by high-affinity zinc-binding proteins in mammalian hosts (29). However, not much work has been done to address this issue. In this line, we have previously shown that A. nidulans and A. fumigatus produce the immunodominant antigens Aspnd1 and Aspf2, respectively, only when the fungi are grown in synthetic media without a zinc supplement (27). These antigens were specifically recognized by human sera obtained from patients with aspergilloma, indicating that these antigens are highly expressed when the fungus grows on living tissue (3). These results led us to investigate the mechanism of zinc uptake in this fungal pathogen. Furthermore, zinc uptake could represent a possible target for an antifungal chemotherapy (5), since A. fumigatus zinc uptake proteins have no significant similarity with proteins exerting a similar function in mammals (20).
To date, microbial zinc uptake has been characterized functionally in detail only in the yeast Saccharomyces cerevisiae. Here, we report the functional characterization of an A. fumigatus zinc uptake system. Our results show that, unlike that observed in S. cerevisiae, the A. fumigatus zinc uptake system depends not only on a low environmental zinc concentration but also on environmental pH. In this work, we focus on the study of the mechanism that operates under acid, zinc-limiting conditions.
MATERIALS AND METHODS
Strains, media, and culture conditions.
All fungal strains used in this work are listed in Table 1. All strains were routinely grown in either PDA complex medium (20 g/liter potato dextrose agar, 20 g/liter sucrose, 2.5 g/liter MgSO4 · 7 H2O) or AMMC medium [10 g/liter dextrose, 0.92 g/liter ammonium tartrate, 0.52 g/liter MgSO4 · 7 H2O, 0.52 g/liter KCl, 1.52 g/liter KH2PO4, 1.1 mg/liter H3BO3, 2.2 mg/liter ZnSO4 · 7 H2O, 0.5 mg/liter MnCl2 · 4 H2O, 0.5 mg/liter FeSO4 · 7H2O, 0.16 mg/liter CoCl2 · H2O, 0.16 mg/liter CuSO4 · 5 H2O, 0.11 mg/liter (NH4)6Mo7O24 · 4 H2O, 5 mg/liter Na2 EDTA, pH adjusted to 6.5 with 1.0 N NaOH] at 37°C for several days. For specific experiments, strains were also grown on SD base medium (5.7 g/liter yeast nitrogen base with ammonium sulfate [without amino acids, dextrose, phosphate, Cu, Zn, or Fe; purchased from Q-BIOgene], 20 g/liter dextrose, 1 g/liter KH2PO4, pH 4.5) which was made a complete SD medium when Fe3+, Cu2+, and Zn2+ were added to SD base medium at a final concentration of 0.74 μM, 0.16 μM, and 100 μM, respectively. Other media that were also used were CDDTE medium (20 g/liter dextrose, 3.0 g/liter NaNO3, 0.5 g/liter MgSO4 · 7 H2O, 0.5 g/liter KCl, 1.0 g/liter KH2PO4, 0.01 g/liter FeSO4 · 7 H2O, 30 mM Trizma base, 250 μM EDTA, pH 8.7) and FBS medium (50% fetal bovine serum, 2% dextrose).
TABLE 1.
Strains used in this study
| Strain | Genotype or description | Reference or source |
|---|---|---|
| A. fumigatus | ||
| CETC | Prototrophic wild type | CECTa |
| CEA17 | Auxotrophic pyrG− | 7 |
| AF14 | Prototrophic wild type (isogenic to CEA17) | This work |
| AF01 | zrfAΔ::neo-pyrG-neo | This work |
| AF02 | zrfAΔ::neo-pyrG-neo | This work |
| AF03 | zrfAΔ::neo-pyrG-neo | This work |
| AF04 | zrfBΔ::hisG-pyrG-hisG | This work |
| AF05 | zrfBΔ::hisG-pyrG-hisG | This work |
| AF13 | zrfBΔ::hisG-pyrG-hisG | This work |
| AF09 | zrfAΔ::neo (auxotrophic pyrG−) | This work |
| AF06 | zrfAΔ::neo zrfBΔ::hisG-pyrG-hisG | This work |
| AF10 | zrfAΔ::neo zrfBΔ::hisG-pyrG-hisG | This work |
| AF11 | zrfAΔ::neo zrfBΔ::hisG-pyrG-hisG | This work |
| AF15 | zrfAΔ::neo zrfBΔ::hisG (auxotrophic pyrG−) | This work |
| AF36 | zrfA-pyrG-neo zrfBΔ::hisG | This work |
| AF37 | zrfA-pyrG-neo zrfBΔ::hisG | This work |
| AF38 | zrfAΔ::neo zrfB-pyrG-hisG | This work |
| AF39 | zrfAΔ::neo zrfB-pyrG-hisG | This work |
| S. cerevisiae | ||
| ZHY3 | MATα ade6 can1 his3 leu2 trp1 ura3 zrt1::LEU2 zrt2::HIS3 | 34 |
Spanish Type Culture Collection.
Metals were added at the concentrations specified for each experiment from sterile solutions of MnCl2 · 4 H2O (20 mM), FeCl2 · 4 H2O (100 mM), FeCl3 · 6 H2O (7.4 mM), CdCl2 · H2O (100 mM), CoCl2 · 6 H2O (100 mM), NiCl2 · 6 H2O (100 mM), CuSO4 · 5 H2O (1.6 mM), or ZnSO4 · 7 H2O (14 mM) in water. All liquid media were sterilized by filtration. Solid media were prepared by adding sterile agar to get a 2% final concentration. In all cases, spores grown in PDA medium were used as inoculum. Prewarmed liquid media were inoculated to a density of 5 × 105 spores/ml and incubated at 37°C with shaking at 200 rpm.
To provide growth conditions at different pH values, a 200 mM PO42−, 400 mM K+ buffer was used supplemented with 200 μM TPEN [N,N,N′,N′tetrakis(2-pyridyl-methyl)ethylenediamine] (P4413; a specific zinc chelator obtained from Sigma). This buffer was added to complete SD medium, without a supplement of zinc, in which A. fumigatus was growing. SD medium was rendered slightly alkaline by including these ions as 57 mM K2HPO4 and 57 μM TPEN (resulting in an initial pH of 7.5); SD was made slightly acid by including 28.5 mM K2HPO4 together with 28.5 mM KH2PO4 plus 28.5 mM KCl and 57 μM TPEN (resulting in an initial pH of 6.5); SD medium was rendered acid by including 57 mM KH2PO4 plus 57 mM KCl and 57 μM TPEN (resulting in an initial pH of 3.5). These pH conditions were formally designated as alkaline, neutral, and acid, respectively. After 1 h of culture at 37°C, pH values did not change.
Yeast strain ZHY3 was routinely grown in yeast extract-peptone-dextrose complex medium at 28°C. For specific experiments, yeast strains were grown on SD medium (5.7 g/liter yeast nitrogen base [obtained from Q-BIOgen], 20 g/liter dextrose, 1 g/liter K2HPO4, 0.74 μM FeCl3 · 6 H2O, and 0.16 μM CuSO4 · 5 H2O) supplemented with 0.67 g/liter of a −LEU −URA dropout mixture (Q-BIOgene). Zinc was added at the final concentration indicated for each particular experiment from a stock solution of ZnSO4 · 7 H2O (14 mM).
DNA manipulations and analyses.
Standard molecular biology procedures for DNA manipulation were used (25). Genomic DNA from A. fumigatus strains was obtained as previously described (3). For Southern blot analyses, 4 μg of DNA per lane was loaded onto a 0.8% agarose gel and transferred by capillarity to positively charged nylon membranes by standard protocols. Probes were digoxigenin labeled by random priming (DIG DNA labeling kit; Roche) and hybridized and detected according to the chemiluminescence method described in the manufacturer's recommendations (DIG nucleic acid detection kit; Roche).
Total RNA was isolated according to the protocol provided with the RNeasy Plant Mini kit (QIAGEN). Twenty micrograms of total RNA per lane was loaded onto 1% formaldehyde agarose gels and transferred by capillarity to neutral nylon membranes according to standard protocols (25). For Northern analysis, fragments of DNA containing most of the open reading frame (ORF) from each gene were labeled by random priming using the DNA Labeling Beads (dCTP) kit and [32P]dCTP (3,000 Ci mmol−1) (Amersham Pharmacia Biotech) and used as probes. Hybridization was carried out at 68°C in a solution containing 0.5 M NaH2PO4 (pH 7.2), 7% sodium dodecyl sulfate (SDS), and 1 mM EDTA. After hybridization, the blots were washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.2% SDS at 68°C for 20 min. The 0.24- to 9.5-kb RNA ladder (Life Technologies) was used for sizing in formaldehyde-agarose gels. As internal RNA load and quality controls, a DNA fragment carrying a partial ORF of the actin gene (acnA) from A. fumigatus was used as a probe.
Isolation of the zrfA and zrfB genes from A. fumigatus.
A DNA fragment from each gene was isolated by a PCR amplification experiment using the degenerate primers JA01 and JA02 (Table 2). These primers were designed according to a pair of amino acid sequences conserved between Zrt1p and Zrt2p of S. cerevisiae (GenBank accession numbers CAA96975 and CAA97701, respectively). These sequences are separated in both cases by 153 and 151 amino acids, respectively. PCR amplification was performed with genomic DNA from the A. fumigatus CECT strain as template. Thirty-five cycles of PCR were performed with the following steps: 45 seconds at 94°C; 1 min at 54°C, and 1.5 min at 72°C. The reaction yielded a fragment with a predicted size of approximately 460 bp. The PCR product was subcloned into pGEM-T easy (Promega). Analysis of several transformants showed that two different plasmids had been obtained: pJAC1 and pJAC2 (Table 2). Sequencing of the PCR fragment carried by each plasmid revealed that two different PCR fragments were in fact amplified. The DNA fragment contained in pJAC1 was similar to a ZRT1 DNA sequence while the one carried by pJAC2 was more similar to a ZRT2 DNA fragment from S. cerevisiae. Next, both PCR products were excised from pJAC1 and JAC2 by EcoRI digestion, digoxigenin labeled by nonradioactive random priming, and used as probes to screen a λEMBL3 A. fumigatus genomic library as previously described (4). The detection of positive clones was done according to the colorimetric method described in the manufacturer's recommendations (DIG nucleic acid detection kit; Boehringer Mannheim). A 3.09-kb XbaI-XbaI DNA fragment containing the entire zrfA gene from A. fumigatus was subcloned into pBlueScript SK(−) to generate pZRF1g. Similarly, a 4.24-kb PvuII-SacI DNA fragment containing the entire zrfB gene was subcloned into pBlueScript SK(−) to generate pZRF24g.
TABLE 2.
Oligonucleotides and plasmids used in this study
| Primer or plasmid | Sequencea or description | Source or reference |
|---|---|---|
| Primers | ||
| JA01 | CTBGARTTYGGHATCATCTTYCA | |
| JA02 | GCCCAYTTDCCGATVARRGCCAT | |
| JA09 | GCACATGAGAGTCGTTTGTGC | |
| JA10 | CCTCGAGTCACCGCTACAACC | |
| JA25 | AACTGAGCGGAATACTCCTCG | |
| JA36 | AAGCTAGCGGCTACCCCTACGACGTCCCCGACTACGCCTTCGACCCCTCCAACGTTGACC | |
| JA37 | CCTGGTTAACGATCATTACGCCCACTTGCC | |
| JA45 | AAGCTAGCGGCTACCCCTACGACGTCCCCGACTACGCCCAAGGCCTTCACACTCTTCTGG | |
| JA46 | GTCAGTTAACCAGAGCGAGATTAGGCCC | |
| RTZ22 | GGGAGTGCAATTATAAGAGTCGCG | |
| Plasmids | ||
| pJAC1 | PCR product (partial sequence of zrfA) | This work |
| pJAC2 | PCR product (partial sequence of zrfB) | This work |
| pMC5-HSET | Carries the ZRT1 gene from S. cerevisiae | 13 |
| pNPN1 | Smaller version of plasmid pCDA14 (7) | This work |
| pNPN2 | Derivative from pNPN1 which carries the pyrG-hisG cassette flanked by sites NsiI-BstZ17I upstream of gene pyrG | This work |
| pPYRG1r | pUC19 derivative carrying the pyrG-hisG cassette flanked by sites EcoRI/BstZ17I upstream of the pyrG gene | This work |
| pPYRG2 | pUC19 derivative carrying the hiG-pyrG-hisG cassette flanked by EcoRI/SmaI/XbaI and HpaI/SpeI sites | This work |
| pRS416 | A centromeric plasmid for expression of genes in S. cerevisiae | 28 |
| pZRF1g | pBluescript carrying a 3.09-kb XbaI fragment containing the zrfA gene | This work |
| pZRF24g | pBluescript carrying a 4.24-kb PvuII-SacI fragment containing the zrfB gene | This work |
| pZHA1 | pMC5-HSET in which the NheI-HpaI fragment had been replaced by the ORF of zrfA | This work |
| pZHA2 | pMC5-HSET in which the NheI-HpaI fragment had been replaced by the ORF of zrfB | This work |
| pZRF1-D4 | pZRF1g in which the BstZ17I-StuI fragment had been replaced by a 6.84-kb HpaI fragment containing the neo-pyrG-neo cassette | This work |
| pZRF2-D6 | pZRF24g in which the XmnI-StuI fragment had been replaced by a 3.88-kb HpaI-SmaI fragment containing the hisG-pyrG-hisG cassette | This work |
| pZRF18-D10 | pNPN2 in which a 2.74-kb NsiI-EcoRV fragment from pZRF1g was ligated into NsiI/BstZ17I | This work |
| pZRF25-D7 | pPYRG1r in which a 3.32-kb EcoRI-PvuII fragment from pZRF24g was ligated into EcoRI/BstZ17I | This work |
Restriction sites are underlined.
Determination of growth rate of A. fumigatus strains.
Dry weight (mg of mycelium per ml of culture) at each time was determined as previously described (2). The generation time (μ) during the log phase (exponential growth) was determined by the formula μ = (tf − t0)/n, where n is the number of generations calculated from the formula n = (logWf − logW0)/log2, in which Wf is the dry weight at the end of the time period (tf) and W0 is the dry weight at the beginning of the time period (t0). The generation times calculated for each strain are the averages from three independent experiments. Using data on generation times, the growth rate (α, h−1) was calculated from the formula α = ln2/μ.
Construction of plasmids used for S. cerevisiae transformation.
The ZHY3 yeast strain was transformed with the centromeric plasmids pZHA1 and pZHA2 (Table 2), which carry the zrfA and zrfB genes, respectively, from A. fumigatus. In these plasmids, the expression of either the zrfA or zrfB ORFs from A. fumigatus is driven by the ZRT1 promoter of S. cerevisiae. To construct these plasmids, only the encoding region of the zrfA and zrfB genes of A. fumigatus was obtained by PCR using plasmid pZRF1g or pZRF24g as a template and the oligonucleotides JA36-JA37 and JA45-JA46, respectively, as primer pairs (Table 2). The quality of PCR products was confirmed by sequencing. The NheI restriction site was included in oligonucleotides JA36 and JA45. The HpaI restriction site was included in oligonucleotides JA37 and JA46. PCR products were digested with NheI and HpaI and used to replace the NheI-HpaI fragment of plasmid pMC5-HSET, which contains nearly the entire ORF of ZRT1 (13). Strain ZHY3 was independently transformed to Ura3+ strains with plasmids pRS416 (28) and pMC5-HSET, which were used as a negative and positive control, respectively. Transformation was carried out according to the LiAc procedure (11). All strains were selected on the appropriate SD dropout medium supplemented with 500 μM ZnSO4 · 7 H2O.
Construction of plasmids used for A. fumigatus transformation.
To obtain a zrfAΔ-null mutant, plasmid pZRF1-D4 was constructed. This plasmid carries a transforming DNA designed to delete most of the ORF of zrfA. To construct this plasmid, a 6.84-kb HpaI-HpaI fragment obtained from plasmid pNPN1, which contains the neo-pyrG-neo cassette, was used to replace a 381-bp BstZ17I-StuI fragment of the ORF from zrfA in plasmid pZRF1g. Similarly, in order to obtain a zrfBΔ-null mutant, a 0.75-kb XmnI-StuI fragment of the ORF of zrfB in plasmid pZRF24g was replaced by a 3.88-kb SmaI-HpaI fragment, which contains the hisG-pyrG-hisG cassette, to generate plasmid pZRF2-D6. The hisG-pyrG-hisG cassette is carried by plasmid pPYRG2, which was constructed at our lab. To obtain revertant strains for either gene zrfA or zrfB, plasmids pZRF18-D10 and pZRF25-D7, respectively, were constructed. A 2.74-kb NsiI-EcoRV fragment from pZRF1g was ligated into NsiI-BstZ17I sites in plasmid pNPN2, which carries the cassette pyrG-neo, to generate plasmid pZRF18-D10. To construct plasmid pZRF25-D7, a 3.32-kb EcoRI-PvuII fragment from pZRF24g was ligated into EcoRI-BstZ17I sites in plasmid pPYRG1r, which carries the cassette pyrG-hisG. Plasmid pZRF2-D4 was linearized by digestion with NheI and NotI, plasmid pZRF2-D6 was linearized by PvuI and NotI digestion, plasmid pZRF18-D10 was linearized by XmnI and NsiI digestion, and plasmid pZRF25-D7 was linearized by digestion with PvuII. In all cases, linearized DNA was extracted with phenol-chloroform-isoamylalcohol, precipitated with isopropanol, resuspended in STC buffer (1 M sorbitol, 10 mM CaCl2, 10 mM Tris-HCl, pH 7.5), and used for transformation as described below.
Protoplast generation and transformation of A. fumigatus.
The protocols used by other laboratories for the preparation of A. fumigatus protoplasts did not work in our hands. However, using such protocols as references, we designed a new procedure which did work in a reproducible fashion. Briefly, conidia were collected in 0.1% Tween 80 from a PDA plate. About 50 ml of liquid AMMC medium supplemented with 0.05% uracil and 0.12% (wt/vol) uridine was inoculated with 8 × 108 spores. The culture was incubated at 28°C for 15 h to induce germination. Mycelia were collected by centrifugation at 3,000 rpm for 10 min at room temperature and washed twice with 0.6 M MgSO4 · 7 H2O. Conidia were resuspended in 10 ml of 1.2 M MgSO4 · 7 H2O. A fresh protoplasting solution was prepared as follows: 1 ml bovine serum albumin (10 mg/ml), 100 mg glucanex (lot no. L1412; Sigma), 0.1 ml zymolyase 20T (50 mg/ml), and 0.2 ml β-glucuronidase (lot no. G8885; Sigma) were dissolved in 7 ml of 1.2 M MgSO4 · 7 H2O, buffered with 80 ml of 1.0 M K2HPO4 and 0.92 ml of 1.0 M KH2PO4, and incubated at room temperature for 5 min. This protoplasting solution was added to the spore suspension and incubated at 32°C for 4 h with gentle shaking. After incubation, the suspension was brought up to 20 ml with 1.2 M MgSO4 · 7 H2O, overlaid with 20 ml of TB medium (0.6 M sorbitol, 0.1 M Tris-HCl, pH 7.5), and centrifuged at 3,000 rpm for 15 min at room temperature in a bucket rotor. Protoplasts were recovered from the interphase and washed once with ST medium (1 M sorbitol, Tris-HCl, pH 7.5) and twice with STC buffer. Protoplasts were resuspended in 0.8 ml of STC buffer. Aliquots of 0.3 ml were used to transform protoplasts of the pyrG− A. fumigatus strain CEA17 using 40 μg of DNA. Next, 75 μl of a 60% polyethylene glycol solution was mixed with the DNA-protoplast mixture and incubated for 20 min on ice. Next, 0.75 ml of additional polyethylene glycol solution was added, mixed, and incubated at room temperature for 20 min. Protoplasts were pelleted and resuspended in 0.2 ml of STC buffer, mixed with 3 ml of soft agar medium, and laid out on an AMMC plate. Plates were incubated at 37°C until pyrG+ fungal transformants had developed. Up to 100 independent transformants for each mutant were reisolated on AMMC medium, and suspensions of conidia from each were prepared in 1.0 ml of water. The correct integration at the expected locus can take place only by means of two recombination events. Those transformants which still bore the gene to be deleted at its original locus were discarded by PCR analysis of genomic DNA obtained from spores as described below. The correct recombination events at the expected loci were verified by Southern blot analyses using the appropriate probes.
Spontaneous pyrG− fungal strains from a pyrG+ independent clone were selected on AMMC medium containing uracil (0.05%), uridine (0.12%), and 5-fluoroorotic acid (1 mg/ml). These pyrG− fungal strains were then used in a second round of transformation to generate a double mutant strain.
Screening of transformants of A. fumigatus by PCR.
Genomic DNA was obtained from spores resuspended in 0.1 ml of 10 mM EDTA and 50 mM Tris-HCl (pH 8.0). Next, 0.1 ml of a denaturing solution (0.2 M NaOH, 1% SDS) was mixed with the spore solution and incubated at 65°C for 30 min. After cooling at room temperature for 10 min, pH was neutralized and SDS was precipitated with 0.1 ml of 3.0 M potassium acetate (pH 5.5). The suspension was cleared by centrifugation for 8 min at 4°C, and the DNA was precipitated with isopropanol, air dried, and resuspended in 10 μl water. The PCR was carried out in a final volume of 25 μl, using 1 μl of DNA suspension as a template. The oligonucleotides used for this purpose must have a GC content up to 60%, preferably either CG or GC at their 3′ end. Oligonucleotides JA09 and JA10 were used in a rapid PCR-based screening in order to discard transformants carrying the neo-pyrG-neo cassette integrated at a locus other than the zrfA locus. The same oligonucleotides were used in a rapid PCR-based screening for positive identification of zrfAΔ pyrG− pop-out strains. Following the same approach, oligonucleotides RTZ22 and JA25 were used to discard transformants carrying the hisG-pyrG-hisG cassette integrated at a locus other than the zrfB locus.
Construction of an isogenic strain of A. fumigatus for use as a control.
As described above, the pyrG− CEA17 strain was used as a recipient for transforming DNA for the construction of A. fumigatus mutants. This strain was generated by chemical mutagenesis, and the mutation at the pyrG locus of this strain has been well characterized at the molecular level (7). However, although in most aspects of its biology this strain has been reported to be similar to the wild-type strain from which it was generated (7), it cannot be precluded that some functions might have been reduced or lost due to additional mutations in loci other than pyrG. Thus, in order to avoid any misinterpretation of the phenotype of the mutants, an isogenic pyrG+ strain of CEA17, called AF14, was constructed following the transformation procedure described above by replacing the damaged pyrG locus of CEA17 with a wild-type DNA carrying the pyrG locus. Revertant pyrG+ strains were selected on AMMC medium without uracil or uridine. In order to ensure that no ectopic integration events had occurred in addition to the expected one, several pyrG+ strains were tested by Southern blot using the same DNA fragment as that used for transformation as a probe.
Nucleotide sequence accession numbers.
The sequence data of the zrfA and zrfB genes have been submitted to the DDBJ, EMBL, and GenBank databases under accession numbers AY611771 and AY611772, respectively.
RESULTS
The products encoded by the zrfA and zrfB genes are membrane zinc transporters.
The zrfA and zrfB genes were identified and cloned as described in Materials and Methods. The zrfA gene has an ORF of 1,077 bp. The initiation transcription site (position +1 in the mRNA) was mapped by rapid amplification of cDNA ends (RACE)-PCR 13 nucleotides upstream from the ATG start codon and 26 nucleotides downstream from the putative TATA box. There are three similar 15-bp sequences in the promoter region that share the same 5′-CAAGGT-3′ core. Preliminary data obtained at our laboratory suggested a role of these sequences in the regulation of zrfA gene expression at the transcriptional level (unpublished data). We have designated these sequences ZRA1, ZRA2, and ZRA3 (for zinc response zrfA), located from positions −60 to −74, −92 to −106, and −142 to −156, respectively (for numbering purposes, we assigned position −1 to the nucleotide preceding the ATG). In addition, from position −94 to −99, located exactly at the middle of the ZRA2 sequence, there is a 5′-GCCARG-3′ PacC-like binding motif (30). Interestingly, no introns were identified within the coding region of zrfA, in contrast to most A. fumigatus genes characterized so far. The ORF of zrfA encodes a 359-amino-acid protein with a predicted molecular mass of 38.9 kDa. Analysis of the amino acid sequence of the ZrfA protein using the Dense Alignment Surface method (6) revealed that it may contain up to eight transmembrane domains. Thus, ZrfA seems to be a membrane-embedded protein that crosses the membrane up to eight times. Transmembrane domains III and IV are separated by a region longer than the rest of the spacer regions. The membrane topology of ZrfA, as predicted by the “positive-inside” rule (31), suggests that both the N and C termini are located on the outer surface of the membrane.
Similarly to zrfA, the zrfB gene has a 1,059-bp ORF not disrupted by introns. It encodes a protein that is structurally similar to ZrfA (353 amino acids and eight transmembrane domains). Interestingly, the initiation transcription site of zrfB mRNA was mapped by RACE-PCR 268 nucleotides upstream from the ATG codon and 28 nucleotides downstream from the putative TATA box. Thus, the 5′ noncoding region of the mRNA is unusually longer than that of most genes from Aspergillus. Also, in the promoter region of zrfB, similarly to the zrfA gene promoter, there are four 15-bp sequences designated ZRB1, ZRB2, ZRB3, and ZRB4, which are located at positions −346 to −360, −481 to −495, −779 to −793, and −878 to −892, respectively. In addition, there are two PacC-like binding motifs extending from positions −282 to −287 and −982 to −987. The identity of ZrfB to ZrfA is 36.6% while similarity increases to 52.8%.
A search in the GenBank database using the BLASTP algorithm revealed that both proteins ZrfA and ZrfB were similar to other putative membrane metal transporters, although they showed the highest similarity to proteins belonging to the Zrt-, Irt-like protein (ZIP) family of transporters involved in cellular zinc uptake (10). Thus, ZrfA showed the highest identity with the EAA66960 protein from Aspergillus nidulans (62.5%), with Zrt1p from S. cerevisiae (54.5%), and with protein EAA33619 from Neurospora crassa (49.0%). ZrfB showed the highest identity with protein EAA60007 from A. nidulans (75.6%), protein EAA52364 from Magnaporthe grisea (42.4%), and Zrt2p from S. cerevisiae (37.6%). Accordingly, while it seems clear that ZrfA may be considered an orthologue of Zrt1p (differences due to speciation phenomena while conserving function), it would not be as easy to consider ZrfB a true orthologue of Zrt2. In fact, ZrfB/Zrt1p identity is higher (35.3%) than that of Zrt1p/Zrt2p (28.7%). Finally, neither ZrfA nor ZrfB shows significant similarities (less than 15%) with any human zinc transporter characterized so far.
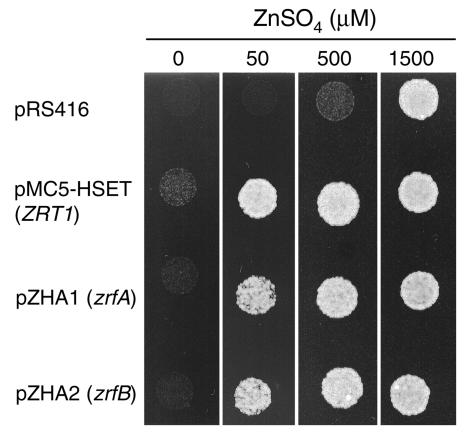
As predicted by structural features and multiple alignment analysis of the ZrfA and ZrfB proteins against many other putative ZIP transporters, the products of the zrfA and zrfB genes may actually encode zinc membrane transporters. To obtain further evidence to support this assumption, the zrfA and zrfB genes were independently expressed in the ZHY3 strain of S. cerevisiae. This yeast strain is not able to grow in severely zinc-limiting conditions unless a high amount of zinc (>1,000 μM) is added to the medium because it lacks the ZRT1 and ZRT2 genes involved in zinc uptake (34). In order to check whether zrfA or zrfB was able to complement the lack of genes involved in zinc uptake in S. cerevisiae, the ORF of either zrfA or zrfB was expressed independently in the ZHY3 strain from a centromeric plasmid constructed as described in Materials and Methods. The ability of ZHY3 transformants to grow under zinc-limiting conditions was tested in complete SD medium (without a supplement of zinc) supplemented with 1 mM EDTA and increasing amounts of zinc (from 0 to 1,500 μM). As shown in Fig. 1, both zrfA and zrfB from A. fumigatus were equally able to complement the lack of the ZRT1 and ZRT2 genes in S. cerevisiae. Thus, this analysis clearly suggested that the proteins encoded by zrfA and zrfB can function as zinc transporters in the background provided by S. cerevisiae, and it supports the idea that the ZrfA and ZrfB proteins may also function as zinc transporters in A. fumigatus.
FIG. 1.
Complementation analysis of zinc uptake provided by the A. fumigatus zrfA and zrfB genes in the ZHY3 yeast strain. This strain was transformed with the empty pRS416 plasmid, plasmid pMC5-HSET (ZRT1), plasmid pZHA1 (zrfA), and plasmid pZHA2 (zrfB). ZHY3 transformed with pRS416 and pMC5-HSET were included as negative and positive controls, respectively. A total of 104 yeast cells were spotted on SD medium supplemented with 1 mM EDTA and increasing amounts of zinc sulfate (0 to 1,500 μM), and plates were incubated for 2 days at 30°C before the picture was taken.
The expression of zrfA and zrfB is regulated in a zinc-dependent fashion.
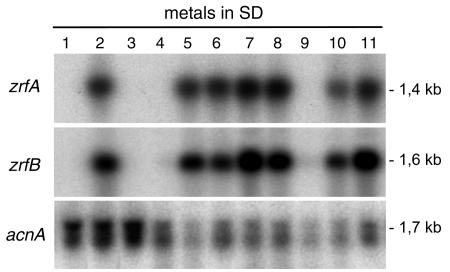
Since zrfA and zrfB encode membrane zinc transporters, and since there are several 15-bp zinc response (ZR) sequences in their promoter regions, we investigated whether the expression of these genes was regulated by zinc. Thus, a wild-type isogenic strain of A. fumigatus (AF14), generated as described in Materials and Methods, was grown for 20 h in complete SD medium and in SD medium without a supplement of either zinc, iron, or copper. Gene transcription was analyzed by Northern blot. In fungal cells grown in complete SD medium, as well as in those grown in SD without a supplement of iron or copper, no transcription of either zrfA or zrfB was detected (Fig. 2). In contrast, a high level of transcription of both genes was observed in fungal cells grown in SD medium without a supplement of zinc. In addition, no other biological metals, such as Fe3+, Cu2+, Mn2+, Fe2+, Co2+, or Ni2+, were able to mimic the repressor effect of zinc on the transcription of both genes, even when SD medium without a supplement of zinc was supplemented with an excess (250 μM) of those metals. Interestingly, only an excess of cadmium, a nonbiological element, was able to mimic the effect of zinc in turning off the transcription of both genes.
FIG. 2.
Northern blot showing the effect of zinc on the transcription of zrfA and zrfB. Strain AF14 was grown in SD medium supplemented as follows: SD medium plus Zn2+ plus Fe3+ plus Cu2+ (1), SD medium minus Zn2+ plus Fe3+ plus Cu2+ (2), SD medium plus Zn2+ minus Fe3+plus Cu2+ (3), SD medium plus Zn2+ plus Fe3+ minus Cu2+ (4), SD medium minus Zn2+ plus Fe3+ plus Cu2+ (5), SD medium minus Zn2+ plus Fe3+ plus Cu2+ (6), SD medium minus Zn2+ plus Fe3+ plus Cu2+ plus Mn2+ (7), SD medium minus Zn2+ plus Fe3+ plus Cu2+ plus Fe2+ (8), SD medium minus Zn2+ plus Fe3+ plus Cu2+ plus Cd2+ (9), SD medium minus Zn2+ plus Fe3+ plus Cu2+ plus Co2+ (10), and SD medium minus Zn2+ plus Fe3+ plus Cu2+ plus Ni2+ (11) (metals in boldface type were added at a final concentration of 250 μM). Probes are indicated to the left of each blot.
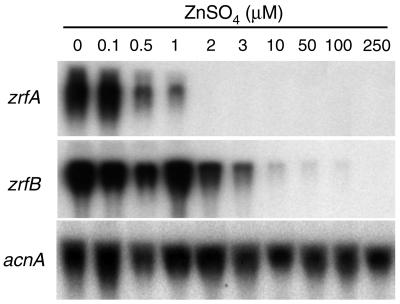
In order to estimate the minimum concentration of zinc initially added to the medium which is required to maintain the transcription of the zrfA and zrfB genes turned off, A. fumigatus (AF14) was grown on SD medium supplemented with increasing amounts of zinc (ranging from 0.0 to 250 μM) for 20 h under the culture conditions described in Materials and Methods, and gene transcription was analyzed by Northern blot (Fig. 3). The initial concentration of zinc required to maintain repressed transcription was 2.0 μM for zrfA and 100 μM for zrfB. This indicates that transcription of zrfA and zrfB is differentially regulated by the environmental concentration of zinc.
FIG. 3.
Repression of transcription of zrfA and zrfB by increasing amounts of zinc. Strain AF14 was grown for 20 h at 37°C in complete SD medium supplemented with zinc sulfate as indicated above each lane. Note that supplement of zinc refers to the amount of zinc initially added to the medium over traces of zinc already present in the medium as a contaminant. However, zinc consumption as a consequence of fungal growth reduced the amount of zinc available in the medium, such that after 20 h of culture, the minimal amount of zinc available in the medium required to repress transcription of each gene is unknown. Probes are indicated to the left of each blot.
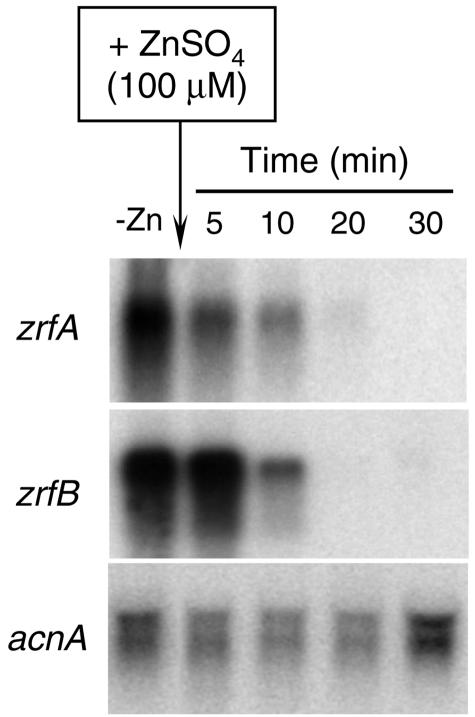
To investigate the stability of the transcriptional product (mRNA) of both genes, A. fumigatus (AF14) was grown for 20 h in SD medium without an initial supplement of zinc to ensure that both genes were being transcribed (Fig. 4). At that time, the culture was supplemented with 100 μM zinc, and total RNA was obtained from samples of mycelia taken after 5, 10, 20, and 30 min of culture. No mRNA for either zrfA or zrfB could be detected after 20 min of incubation as observed by Northern blot which shows that mRNA is being degraded rapidly as soon as the transcription of new mRNA molecules stops. Interestingly, identical results were obtained when repression was triggered with a concentration of zinc in the medium as low as 1 μM (data not shown), which suggests that the transcriptional mechanism which controls transcription of either zrfA or zrfB is sensible to any sudden slight increase of zinc in the environment. In sum, these results suggest that the control of the transcription of the zrfA and zrfB genes must lie in the first stage of a tightly regulated mechanism involved in zinc uptake.
FIG. 4.
Life span (turnover) of zrfA and zrfB mRNA after repression of transcription by zinc. Strain AF14 was grown in complete SD medium without a supplement of zinc for 20 h, and a sample was taken (lane 1, −Zn). At this point, the culture was supplemented with 100 μM zinc and samples were collected and immediately frozen in liquid nitrogen after 5, 10, 20, and 30 min. Probes are indicated to the left of each blot.
zrfA and zrfB are required for optimal growth of A. fumigatus under zinc-limiting conditions.
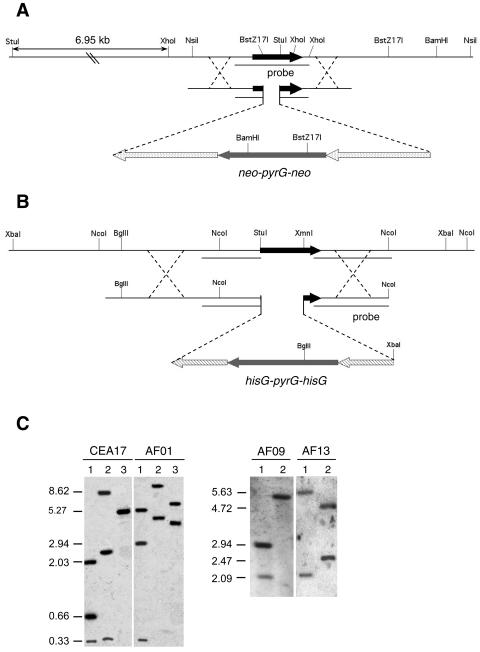
In order to confirm the function of these genes in A. fumigatus, a mutant strain for each gene and a double zrfAzrfBΔ mutant strain were constructed by the gene replacement technique described in Materials and Methods. Briefly, a 381-bp fragment of the zrfA-encoding region was replaced by the neo-pyrG-neo cassette in the CEA17 pyrG− strain to generate strains AF01, AF02, and AF03 (Fig. 5A). Similarly, a 750-bp fragment containing almost the entire open reading frame of the zrfB gene was replaced by the hisG-pyrG-hisG cassette in strain CEA17 to generate strains AF04, AF05, and AF13 (Fig. 5B). The AF06, AF10, and AF11 double zrfAzrfBΔ mutant strains were constructed by replacing the zrfB gene by the hisG-pyrG-hisG cassette in strain AF09, a spontaneous zrfAΔ pyrG− strain obtained from AF01 as described in Materials and Methods. The recombination events occurring in the AF09 strain were identical to those taking place when the zrfB gene was replaced in strain CEA17. All strains harbored the correct integration event at the expected locus, as verified by Southern blot analyses (Fig. 5C). In order to more clearly assess the function of each gene over the growth of A. fumigatus, and following a strategy similar to that used to construct either zrfAΔ- or zrfBΔ-null mutants, a revertant strain for each gene was generated after reintroducing independently a wild-type copy of either gene zrfA or zrfB at its original locus into the strain AF15, a spontaneous zrfAzrfBΔ pyrG− strain obtained from AF10 (data not shown). The gene zrfA was reintroduced in AF15 using the zrfA-pyrG-neo cassette to generate strains AF36 and AF37, two independent zrfBΔ mutant strains that should growth identically to any single zrfBΔ mutant strain. The gene zrfB was reintroduced into AF15 using the zrfB-pyrG-hisG cassette to generate strains AF38 and AF39, two independent zrfAΔ mutant strains that should present growth identical to that of any single zrfAΔ mutant strain.
FIG. 5.
(A) Schematic representation of the integration of the neo-pyrG-neo cassette used to disrupt the zrfA gene by a deletion-substitution mutation. (B) Schematic representation of the integration of the hisG-pyrG-hisG cassette used to disrupt the zrfB gene by a deletion-substitution mutation. Only relevant restriction sites have been indicated in both schemes. (C) Southern blots confirming that the integration event had taken place as expected. Genomic DNA from strains CEA17 (as a control) and AF01 was digested with XhoI/BstZ17I (lane 1), StuI/BstZ17I (lane 2), and NsiI/BamHI (lane 3) and hybridized with the probe indicated in panel A. Similarly, genomic DNA from strains AF09 (as a control) and AF13 was digested with NcoI (lane 1) and XbaI/BglII (lane 2) and was hybridized with the probes indicated in panel B. The exact sizes (in kb) of the hybridizing DNA fragments are indicated to the left of each blot.
Phenotypic analysis was performed for all the pyrG+ mutant strains. The three independent zrfAΔ mutant strains (AF01, AF02, and AF03) grow identically under all conditions tested as described below, as do the three independent zrfBΔ mutant strains (AF04, AF05, and AF13). Similarly, the three independent zrfAzrfBΔ mutant strains (AF06, AF10, and AF11) as well as the two independent zrfA revertant strains (AF36 and AF37) and the two independent zrfB revertant strains (AF38 and AF39) also grew identically under all conditions tested. Thus, for simplicity, only mutant strains AF01 (zrfAΔ), AF13 (zrfBΔ), AF10 (zrfAzrfBΔ), AF36, and AF39 will be mentioned from now on.
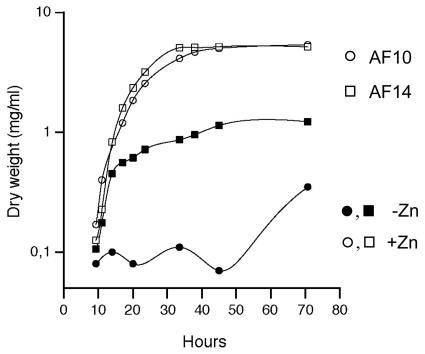
Strains AF01, AF13, and AF10 of A. fumigatus and the wild-type AF14 isogenic strain were grown in SD liquid medium under both zinc-replete and zinc-limiting conditions (Fig. 6 and Table 3). All strains showed similar growth rates (calculated as described in Materials and Methods) and reached similar maximum dry weights when they were grown under zinc-replete conditions (SD medium plus 100 μM zinc). In contrast, under zinc-limiting conditions (SD medium without a supplement of zinc), strains AF14, AF01, and AF13, as well as revertant strains AF36 and AF39, equally reduced (about fourfold) their maximum dry weight, while strain AF10 reduced it by 16-fold. Similarly, under zinc-limiting conditions, the growth rates of strains AF14, AF01, AF13, AF36, and AF39 were similar, although slightly reduced (about 1.1-fold), in comparison with growth in the presence of zinc. However, the growth rate of strain AF10 under zinc-depleted conditions was so low that it could not be measured accurately. This indicates that either the zrfA or zrfB gene is required for optimal growth of A. fumigatus in liquid medium under zinc-limiting conditions.
FIG. 6.
For clarity, only the growth curves of the isogenic AF14 wild type (squares) and the double mutant AF10 zrfAzrfBΔ strain (circles) are represented. These strains were grown in complete SD medium supplemented with 100 μM zinc sulfate (open symbols) or without a zinc supplement (closed symbols) for 70 h at 37°C.
TABLE 3.
Growth data for A. fumigatus strains in liquid SD medium
| Strain | α (1/h)
|
Max. dry wt (mg/ml)a
|
||
|---|---|---|---|---|
| SD + Zn | SD − Zn | SD + Zn | SD − Zn | |
| AF14 | 0.33 ± 0.04 | 0.30 ± 0.03 | 5.18 ± 0.2 | 1.22 ± 0.2 |
| AF01 | 0.31 ± 0.03 | 0.27 ± 0.01 | 5.07 ± 0.3 | 1.23 ± 0.1 |
| AF13 | 0.33 ± 0.07 | 0.27 ± 0.05 | 5.51 ± 0.1 | 1.47 ± 0.1 |
| AF10 | 0.30 ± 0.01 | NDb | 5.28 ± 0.2 | 0.32 ± 0.1 |
| AF36 | 0.32 ± 0.02 | 0.28 ± 0.01 | 5.20 ± 0.1 | 1.25 ± 0.1 |
| AF39 | 0.30 ± 0.02 | 0.28 ± 0.02 | 5.20 ± 0.1 | 1.22 ± 0.1 |
Determined after 65 h of culture in liquid SD medium with or without a supplement of zinc.
ND, not determined.
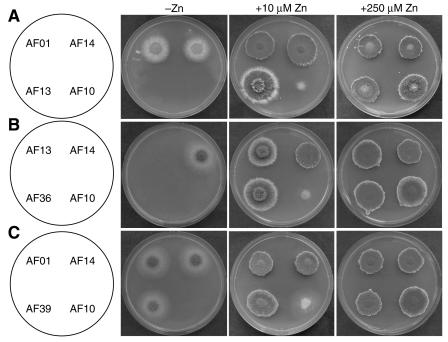
To test the function of the zrfA and zrfB genes on the growth of A. fumigatus in solid medium, all strains were cultured in SD agar, which was converted to a severely zinc-limiting medium by adding 250 μM EDTA (SDE medium). In this medium, maximal growth was reached after 6 days of incubation regardless of the zinc supplement added to medium, although normal growth in all strains (including the wild type) could be achieved only at a concentration of zinc of 250 μM or higher in the medium (Fig. 7A). Interestingly, while the wild-type (AF14) and zrfAΔ (AF01) strains grew identically in SDE medium supplemented with 10 μM zinc, strain zrfBΔ (AF13) grew generating cottony colonies formed by narrow but long extended and low branched hyphae in which a delay in sporulation was observed at the edge of the colony. Interestingly, this pattern of growth resembles that of the AF14 wild-type strain grown on solid SDE medium without a supplement of zinc. The zrfAzrfBΔ (AF10) strain was unable to grow. In addition, a zrfBΔ strain in which a wild-type copy of the gene zrfA was reintroduced at its original locus (AF36) grew identically to strain AF13 (zrfBΔ) (Fig. 7B). Similarly, a zrfAΔ strain in which a wild-type copy of the gene zrfB has been reintroduced at its original locus (AF39) grew identically to strain AF01 (zrfAΔ) (Fig. 7C). Although the growth defects observed in strains AF13, AF10, and AF36 are probably due to the sequestering of zinc by EDTA, it is still possible that some other essential metal ion could also be sequestered by EDTA, lowering its concentration to a suboptimal level for the normal growth of these strains. Thus, to preclude this possibility, we tested whether the SDE medium with the addition of 10 μM zinc supplemented with other metals (Cu2+, Fe3+, Co2+, Ni2+, or Mn2+) at the same molar rate as EDTA (i.e., 250 μM) could improve the growth of strains AF13, AF10, and AF36 to a similar extent to that of either the AF14 or the AF01 strain. However, only zinc allowed all strains to reach their maximal growth (data not shown), confirming the notion that the alteration of growth in AF13, AF10, and AF36 would be due to a loss of function of these mutants in taking up zinc from the medium.
FIG. 7.
(A) Growth of the wild-type strain and mutants in SDE zinc-limiting medium without a supplement of zinc or with a supplement of either 10 or 250 μM zinc sulfate. (B) Comparison of growth of the zrfA revertant strain AF36 to that of the zrfBΔ mutant strain AF13 in the same medium used in the upper panel. (C) Comparison of growth of the zrfB revertant strain AF39 to that of the zrfAΔ mutant strain AF10 in the same medium used in the upper panel. In all cases, 103 spores of each strain were spotted onto the same plate and arranged as indicated in the scheme, and the inoculated plates were incubated for 6 days at 37°C in a humid atmosphere before the picture was taken.
In sum, the analysis of growth of the mutants in SDE medium may be interpreted as that the expression of zrfA only is required for A. fumigatus to grow (at a suboptimal level) in a zinc-limiting medium in the absence of zrfB. However, if it is taken into account that (i) either zrfA or zrfB encodes proteins required for optimal growth under zinc-limited conditions as shown by the yeast complementation experiments (Fig. 1), (ii) the expression of either zrfA or zrfB is detected by Northern blot in zinc-limiting SD medium (Fig. 3), and (iii) the zrfBΔ strain (AF13) does not grow as well as the zrfAΔ strain (AF01) or the wild-type strain (AF14) (Fig. 7A), we should conclude that both genes are actually required for optimal growth and differentiation of the fungus in solid medium in which zinc is found in limited amounts, even though zrfA makes a smaller contribution.
zrfA and zrfB are acid-expressed genes in zinc-limiting conditions.
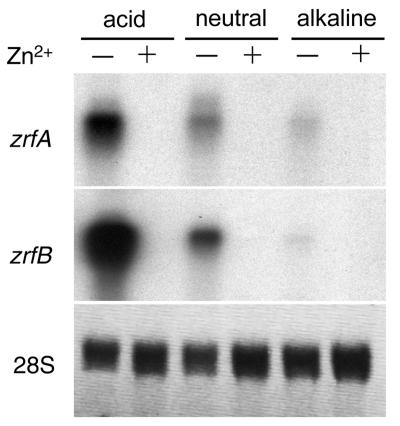
Just as a relationship was seen between the existence of ZR motifs in the promoter regions of the zrfA and zrfB genes and their expression, depending on the environmental concentration of zinc, a relationship may also exist between the existence of PacC-like sequences in their promoters and the regulation of their expression in a pH-dependent manner at the transcriptional level. In addition, the apparently strategic localization of PacC-like sequences in the promoter regions of both zrfA and zrfB (a PaC-like sequence in the zrfA promoter is located exactly at the middle of its ZRA2 sequence, while in the zrfB promoter, a PacC-like sequence is located just downstream from its TATA box) led us to suspect that the transcription of these genes might be regulated in response to two different environmental stimuli, such as the concentration of zinc and pH. To address this issue, the AF14 wild-type strain was grown in SD medium without a supplement of zinc for 24 h in order to enable the expression of both genes zrfA and zrfB, after which the culture was split into three aliquots that were buffered at either acid pH (3.5), neutral pH (6.5), or alkaline pH (7.5), as described in Materials and Methods. Next, each buffered aliquot of the culture was divided in two flasks, one of them supplemented with an excess of zinc (200 μM), while the other was not. Analysis of gene expression under these new conditions revealed that transcription of both genes was progressively repressed as the pH of the medium turned alkaline. Transcription of zrfA and zrfB in neutral or alkaline medium was equally reduced but only under zinc-limiting conditions, since in zinc-replete media, the expression of both genes was repressed regardless of pH (Fig. 8). These results clearly showed that the zrfA and zrfB genes are acid-expressed genes under zinc-limiting conditions. In addition, these results suggest that for these genes, the regulatory mechanism of gene expression that controls zinc availability prevails over the one that responds to changes in the environmental pH. Thus, pH-dependent regulation must play a secondary role in the transcriptional expression of both genes.
FIG. 8.
Regulation of the expression of the zrfA and zrfB genes at the transcriptional level in response to changes in environmental pH under either zinc-replete (SD medium plus 200 μM Zn2+) or zinc-limiting conditions. Probes are indicated to the left of each blot.
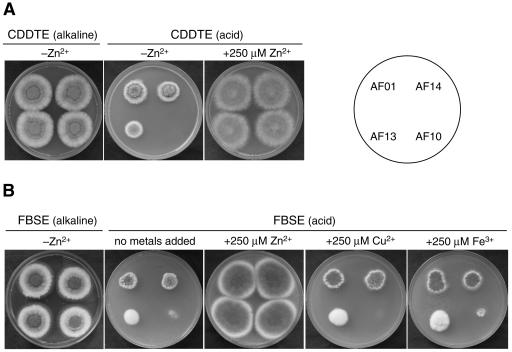
To further investigate the relationship between the expression of the zrfA and zrfB genes and the growth of A. fumigatus depending on the environmental pH, all strains were grown in alkaline CDDTE (pH 8.7) and in acid CDDTE (pH was lowered to 4.5 with HCl) agar media (Fig. 9A). The CDDTE medium is an extremely zinc-limiting medium since it does not have a supplement of zinc and contains 250 μM EDTA. All fungal strains grew equally well in alkaline CDDTE medium without a supplement of zinc. In contrast, in acid CDDTE medium without a supplement of zinc, the zrfBΔ AF13 strain showed a slightly reduced growth capacity, while the zrfAzrfBΔ AF10 strain did not grow at all. Interestingly, an excess of zinc (250 μM) improved the growth of strains AF13 and AF10 to a level similar to that seen for AF01 and AF14 strains, while an excess amount of other metals, such as copper or iron, was completely unable to improve the growth of either the AF13 or AF10 strain (data not shown). This shows that, as for growth in acid SDE medium (Fig. 8), and although we cannot appreciate a defect in the growth of the AF01 zrfAΔ strain culture on acid, zinc-limiting CDDTE medium, the optimal growth of the fungus under this conditions can be achieved only when expression of both genes occurs.
FIG. 9.
Effect of zinc and pH on growth of A. fumigatus strains. (A) Strains were grown in alkaline CDDTE medium (without a supplement of zinc), acid CDDTE medium (without a supplement of zinc), and acid CDDTE medium supplemented with 250 μM zinc. (B) Strains were grown in FBSE medium (pH 7.5) without a supplement of zinc, in acid FBSE medium (pH 4.5) without a supplement of any metal, and in acid FBSE medium supplemented with 250 μM zinc sulfate, copper sulfate, or ferric chloride. In all cases, 103 spores of each strain, arranged as indicated in the scheme, were spotted onto the same plate. Plates were incubated for 3 days at 37°C in a humid atmosphere before the picture was taken.
Since A. fumigatus is an opportunistic pathogen, we also tested the growth of these mutants in fetal bovine serum (FBS) agar. FBS has a neutral or slightly alkaline pH (pH 7.2 to 7.5) that does not decline during incubation and remains alkaline (12). In addition, even though metal cations do not remain soluble in water as the pH becomes neutral or alkaline, metals do not precipitate since the main pool of each metal is usually tightly bound to chelating proteins present in the serum. FBS was also supplemented with 250 μM EDTA (FBSE) (pH 7.5) to chelate traces of free zinc and to render the medium more zinc limiting. All A. fumigatus strains grew healthily in FBSE without a supplement of zinc (Fig. 9B). In contrast, when FBSE was acidified with HCl to a pH value of 4.5, strains AF14, AF01, and AF13 were able to grow (strain AF13 grew more slowly), while AF10 did not grow at all. However, when acid FBSE was supplemented with zinc, all strains improved their growth to a level identical to that of the wild type, including strain AF10, which regained its full growth ability. In addition, it was observed that the inability of the AF10 strain to grow in acidified FBSE was specifically linked to a defect in zinc uptake under zinc-limiting conditions, since all strains improved their capacity to grow only when they were supplemented with an excess amount of zinc but not with similar amounts of either copper or iron. Revertant strains AF36 and AF39 grew identically to strains AF13 and AF01, respectively, in either CDDTE or FBSE medium under all growth conditions tested (data not shown). These results show that the zrfA and zrfB genes are irrelevant for zinc uptake and growth in normal serum even under zinc-limiting conditions.
In summary, the zrfA and zrfB genes are required for optimal growth under acid, zinc-limiting conditions, but they are not apparently required for growth under alkaline, zinc-limiting conditions. In addition, provided that pH and zinc depletion conditions are maintained, the patterns of zrfA and zrfB gene expression do not depend on the medium used, since growth was identical in media as different as CDDTE medium, a synthetic defined medium, and fetal bovine serum, a complex medium that somehow resembles the environment that may find the fungus when it grows in living tissues.
DISCUSSION
We reported for the first time that the in vitro expression of immunodominant antigens from Aspergillus spp. is up-regulated by a low environmental concentration of zinc (27). This result prompted us to conjecture that the acquisition of zinc by A. fumigatus from a zinc-limiting environment, such as a living animal, must be one of the main difficulties that this fungal pathogen must overcome to survive and grow under these conditions. Thus, based on insight gained from the investigation of the mechanisms of zinc uptake in S. cerevisiae (33, 34), we assessed the role of zinc uptake in the growth of A. fumigatus. First, we identified two genes in A. fumigatus (zrfA and zrfB) that encoded proteins ZrfA and ZrfB, which are structurally similar to proteins Zrt1p and Zrt2p, respectively, from S. cerevisiae. All these proteins belong to the ZIP family of zinc transporters (10). In the promoter regions of the A. fumigatus genes, there were several 15-bp ZR sequences that are probably involved in their regulation at the transcriptional level, as suggested by preliminary data obtained in our laboratory. Interestingly, the core sequence found in all A. fumigatus 15-bp ZR sequences is also found at the 3′ end of the zinc response elements identified in yeast promoters whose expression is somehow regulated by zinc (32). In fact, we observed that only a high concentration of zinc down-regulated their expression and that no metal other than Cd2+ affected the transcriptional expression of these genes. Interestingly, it has repeatedly been reported that Cd2+ mimics Zn2+ in its ability to bind biological substrates (21, 24), probably due to the very similar electronic conformation that exists between the two elements. In any case, cadmium is a toxic element, since even though it can replace Zn2+ structurally, it cannot replace it functionally (16). In addition, we also observed that the expression of zrfA and zrfB was differentially down-regulated at the transcriptional level by concentrations of environmental zinc as low as 2 to 10 μM. Transcription of zrfB was totally turned off at a concentration of zinc approximately 50-fold higher than that required to repress transcription of zrfA. Interestingly, a similar mechanism of differential transcriptional regulation which depends on the environmental zinc concentration has also been described for the ZRT1 and ZRT2 genes from S. cerevisiae. A zrt1 mutant strain of S. cerevisiae showed a marked reduction in growth, while a zrt2 mutant grew as a wild-type yeast strain under zinc-limiting conditions. This can be explained in terms of the notion that the ZRT1 and ZRT2 genes encode the high- and low-affinity zinc uptake systems, respectively, of S. cerevisiae (33, 34). Similarly to S. cerevisiae strains deficient in zinc uptake, the growth (i.e., maximal dry weight) of a zrfAzrfBΔ A. fumigatus mutant was reduced by fourfold compared to that of a wild-type strain. However, even though the ZrfA and ZrfB proteins of A. fumigatus seem to be orthologues of Zrt1p and Zrt2p from S. cerevisiae, respectively, the growth of A. fumigatus zrfAΔ mutants was nearly identical to that of the wild type, whereas the growth of zrfBΔ mutants had an effect on the growth of A. fumigatus that strongly resembled that of the zrt1 mutation on the growth of S. cerevisiae. This suggests one of the following possibilities. First, despite the overall similarity between Zrt1p/ZrfA and Zrt2p/ZrfB, A. fumigatus proteins would not be true orthologues of the yeast proteins, such that ZrfA and ZrfB would be responsible for a low- and high-affinity zinc uptake system, respectively. Second, ZrfA and ZrfB could be true orthologues of yeast proteins, such that they would show high- and low-affinity zinc transport activity, respectively, but a higher level of expression of zrfB might compensate for its low zinc transport activity, making it the most relevant system operating in zinc uptake in A. fumigatus. Third, both ZrfA and ZrfB would be transporters with a similar zinc transport activity, which would be closer to that exhibited by Zrt1p. This last assumption is supported by the fact that both genes improved the growth of the ZHY3 yeast strain to an equal extent under severe zinc-limiting conditions (SD medium plus 1 mM EDTA plus 50 μM Zn2+). In any case, since the transcription of zrfB was repressed at a higher concentration of zinc than the transcription of zrfA, the expression of zrfB may be the main system for acquisition of zinc in A. fumigatus cells under a broader range of environmental zinc concentrations.
The existence of PacC-like sequences in the promoter region of the zrfA and zrfB genes prompted us to test whether the transcriptional expression of zrfA and zrfB might somehow also be regulated by the ambient pH. In addition, since the PacC-like sequence of the zrfA promoter overlaps the 15-bp ZR sequence, alkaline pH may directly repress the transcription of zrfA by blocking transcriptional induction as a result of a competitive binding mechanism as suggested previously by other investigators (9). However, it is also possible that at alkaline pH, the binding of the PacC active form to the binding site located just 9 nucleotides downstream from the zrfB TATA box might interfere with the efficient binding of RNA polymerase and the transcription of zrfB. Whichever mechanism is in fact operating, the transcription of these genes will be up-regulated in an acid environment, whereas it will be down-regulated in an alkaline, zinc-limiting environment. On the other hand, since the transcription of these genes is also regulated by pH, it is still possible that the ZrfA and ZrfB proteins may function when a zinc-limiting environment turns acid. This is unlikely, however, because S. cerevisiae strains expressing the A. fumigatus genes grew identically at both acid and alkaline pH (data not shown), suggesting that the ZrfA and ZrfB proteins in the yeast background function in a pH-independent manner.
Serum stimulates the growth of A. fumigatus, even though serum contains metal-binding proteins that sequester free-ion metals, affording a concentration of free-ion metals too low to support the growth of many microorganisms, including most pathogenic fungi (12). Thus, pathogens must have developed mechanisms to obtain metals from serum. One of these mechanisms that has been developed by A. fumigatus, as well as by other pathogens, is the production of siderophores to scavenge iron from serum (17). However, this system would not be efficient unless an iron uptake system were coupled to it. In fact, specific transporters for siderophores have recently been characterized in A. nidulans, which indicates that orthologues probably exist in A. fumigatus (8, 14, 15). Likewise, even though no system has been described for zinc scavenging to date, a zinc uptake system would be required by pathogens, including A. fumigatus, to incorporate zinc into cells. The results obtained by us show that zrfA and zrfB are important genes for zinc uptake but only under acidic conditions. Accordingly, another system must exist in addition to this one, which would be used by fungi to obtain zinc under alkaline conditions. In fact, we have identified another zinc transporter-encoding gene (named as zrfC) that is expressed only under alkaline conditions (unpublished data). This system as well as the transcriptional regulatory mechanism that underlies the expression of all zinc-expressed genes are currently under active investigation at our laboratory.
Acknowledgments
We express our gratitude to C. d'Enfert (Institut Pasteur) and D. Eide (University of Columbia, Columbia, MO) for kindly providing the plasmids and strains.
This work was supported by grants SA039/01 and BIO2001-1692 from the Junta de Castilla y León and the Spanish Ministerio de Ciencia y Tecnología, respectively. R.V. and M.Á.M. were recipients of fellowships from the Ministerio de Ciencia y Tecnología (Spain).
REFERENCES
- 1.Auld, D. S. 2001. Zinc coordination sphere in biochemical zinc sites. BioMetals 14:271-313. [DOI] [PubMed] [Google Scholar]
- 2.Calera, J. A., R. López-Medrano, M. C. Ovejero, P. Puente, and F. Leal. 1994. Variability of Aspergillus nidulans antigens with media and time and temperature of growth. Infect. Immun. 62:2322-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calera, J. A., M. C. Ovejero, R. López-Medrano, M. Segurado, P. Puente, and F. Leal. 1997. Characterization of the Aspergillus nidulans aspnd1 gene demonstrates that the ASPND1 antigen, which it encodes, and several Aspergillus fumigatus immunodominant antigens belong to the same family. Infect. Immun. 65:1335-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calera, J. A., S. Paris, M. Monod, A. J. Hamilton, J. P. Debeaupuis, M. Diaquin, R. López-Medrano, F. Leal, and J. P. Latge. 1997. Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus. Infect. Immun. 65:4718-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chimienti, F., M. Aouffen, A. Favier, and M. Seve. 2003. Zinc homeostasis-regulating proteins: new drug targets for triggering cell fate. Curr. Drug Targets 4:323-338. [DOI] [PubMed] [Google Scholar]
- 6.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in procariotic membrane proteins: the Dense Alignment Surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 7.d'Enfert, C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76-82. [DOI] [PubMed] [Google Scholar]
- 8.Eisendle, M., H. Oberegger, I. Zadra, and H. Haas. 2003. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding L-ornithine N5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol. Microbiol. 49:359-375. [DOI] [PubMed] [Google Scholar]
- 9.Espeso, E. A., and H. N. Arst, Jr. 2000. On the mechanism by which alkaline pH prevents expression of an acid-expressed gene. Mol. Cell. Biol. 20:3355-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaither, L. A., and D. Eide. 2001. Eukaryotic zinc transporters and their regulation. BioMetals 14:251-270. [DOI] [PubMed] [Google Scholar]
- 11.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 12.Gifford, A. H. T., J. R. Klippenstein, and M. M. Moore. 2002. Serum stimulates growth of and proteinase secretion by Aspergillus fumigatus. Infect. Immun. 70:19-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gitan, R. S., H. Luo, J. Rodgers, M. Broderius, and D. Eide. 1998. Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 273:28617-28624. [DOI] [PubMed] [Google Scholar]
- 14.Haas, H. 2003. Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl. Microbiol. Biotechnol. 62:316-330. [DOI] [PubMed] [Google Scholar]
- 15.Haas, H., M. Schoeser, E. Lesuisse, J. F. Ernst, W. Parson, B. Abt, G. Winkelmann, and H. Oberegger. 2003. Characterization of the Aspergillus nidulans transporters for the siderophores enterobactin and triacetylfusarinine C. Biochem. J. 371:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartwig, A. 2001. Zinc finger proteins as potential targets for toxic metals ions: differential effects on structure and function. Antioxid. Redox Signal 3:625-634. [DOI] [PubMed] [Google Scholar]
- 17.Hissen, A. H. T., J. M. T. Chow, L. J. Pinto, and M. M. Moore. 2004. Survival of Aspergillus fumigatus in serum involves removal of iron from transferrin: the role of siderophores. Infect. Immun. 72:1402-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard, D. H. 1999. Acquisition, transport, and storage of iron by pathogenic fungi. Clin. Microbiol. Rev. 12:394-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibrahim-Granet, O., B. Philippe, H. Boleti, E. Boisvieux-Ulrich, D. Grenet, M. Stern, and J. P. Latge. 2003. Phagocytosis and intracellular fate of Aspergillus fumigatus conidia in alveolar macrophages. Infect. Immun. 71:891-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kambe, T., Y. Yamaguchi-Iwai, R. Sasaki, and M. Nagao. 2004. Overview of mammalian zinc transporters. Cell. Mol. Life Sci. 61:49-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamizono, A., M. Nishizawa, Y. Teranishi, K. Murata, and A. Kimura. 1989. Identification of a gene conferring resistance to zinc and cadmium ions in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 219:161-167. [DOI] [PubMed] [Google Scholar]
- 22.Kontoyiannis, D. P., and G. P. Bodey. 2002. Invasive aspergillosis in 2002: an update. Eur. J. Clin. Microbiol. Infect. Dis. 21:161-172. [DOI] [PubMed] [Google Scholar]
- 23.Nilius, A. M., and S. G. Farmer. 1990. Identification of extracellular siderophores of pathogenic strains of Aspergillus fumigatus. J. Med. Vet. Mycol. 28:395-403. [DOI] [PubMed] [Google Scholar]
- 24.Palumaa, P., O. Njunkova, L. Pokras, E. Eriste, H. Jörnvall, and R. Sillard. 2002. Evidence for non-isostructural replacement of Zn2+ with Cd2+ in the β-domain of brain-specific metallothionein-3. FEBS Lett. 527:76-80. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schrettl, M., E. Bignell, C. Kragl, C. Joechl, T. Rogers, H. J. Arst, K. Haynes, and H. Haas. 2004. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segurado, M., R. López-Aragon, J. A. Calera, J. M. Fernández-Ábalos, and F. Leal. 1999. Zinc-regulated biosynthesis of immunodominant antigens from Aspergillus spp. Infect. Immun. 67:2377-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugarman, B. 1983. Zinc and infection. Rev. Infect. Dis. 5:137-147. [DOI] [PubMed] [Google Scholar]
- 30.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Peñalva, and H. N. Arst, Jr. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Heijne, G. 1992. Membrane protein structure prediction hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 32.Zhao, H., E. Butler, J. Rodgers, T. Spizzo, S. Duesterhoeft, and D. Eide. 1998. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem. 273:28713-28720. [DOI] [PubMed] [Google Scholar]
- 33.Zhao, H., and D. Eide. 1996. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93:2454-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao, H., and D. Eide. 1996. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271:23203-23210. [DOI] [PubMed] [Google Scholar]