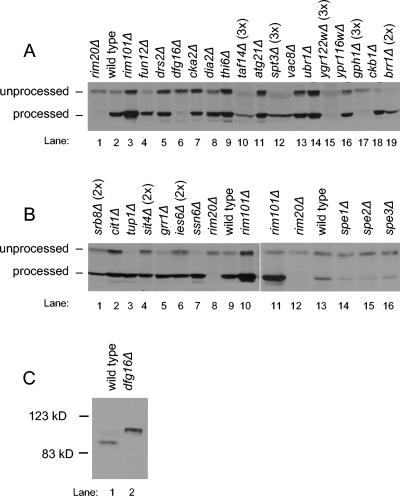
FIG. 1.
Processing of Ura3-V5-Rim101p and Rim101-HA2p. (A, B) Protein extracts from MATa deletion library S. cerevisiae strains containing a URA3-V5-RIM101 plasmid were analyzed on an anti-V5 immunoblot to visualize the unprocessed and processed forms of the protein. Protein amounts loaded were approximately equal, as determined by Ponceau-S staining, with the exception of panel A, lanes 10, 12, 15, 17, and 19, and panel B, lanes 1, 4, and 6. These lanes were intentionally loaded with two or three times as much protein, as indicated above the lane, in order to detect the epitope. (C) Protein extracts from yeast strains WXY169 (RIM101-HA2 DFG16) and YKB167 (RIM101-HA2 dfg16Δ) were analyzed on an anti-HA immunoblot to visualize the processing of the epitope-tagged Rim101p, expressed from the native RIM101 locus.