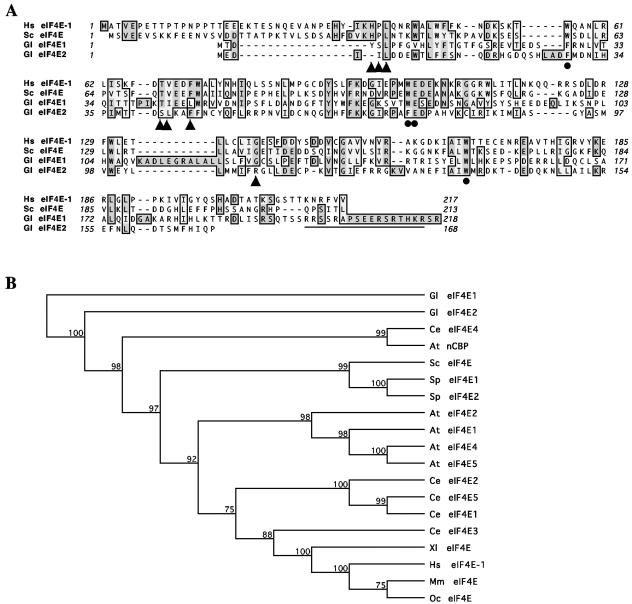
FIG. 1.
Alignment and phylogeny of eIF4Es from Giardia and other organisms. (A) Alignment of Giardia eIF4Es with human eIF4E-1 and S. cerevisiae eIF4E. The predicted amino acid sequences of Giardia eIF4E1 and eIF4E2 were aligned with those of human eIF4E-1 and S. cerevisiae eIF4E using the CLUSTAL W algorithm (47). Identical residues are shaded in dark gray, and related residues are in light gray. Residues with dots underneath are essential for cap binding, whereas triangles indicate residues essential for interaction with eIF4G in humans and yeast (27). The sequences predicted to be the bipartite nucleus localization signal in Giardia eIF4E1 is underlined. (B) Phylogeny of eIF4Es. The selected eIF4Es were analyzed using unweighted-pair group method using average linkages distance matrix methods from the MacVector software. The bootstrap tree was shown with 1,000 distance replicates performed. Abbreviated sequence identifiers are as follows: At, A. thaliana; Ce, C. elegans; Gl, G. lamblia; Hs, Homo sapiens; Mm, Mus musculus; Sc, S. cerevisiae; Sp, S. pombe; Xl, Xenopus laevis.