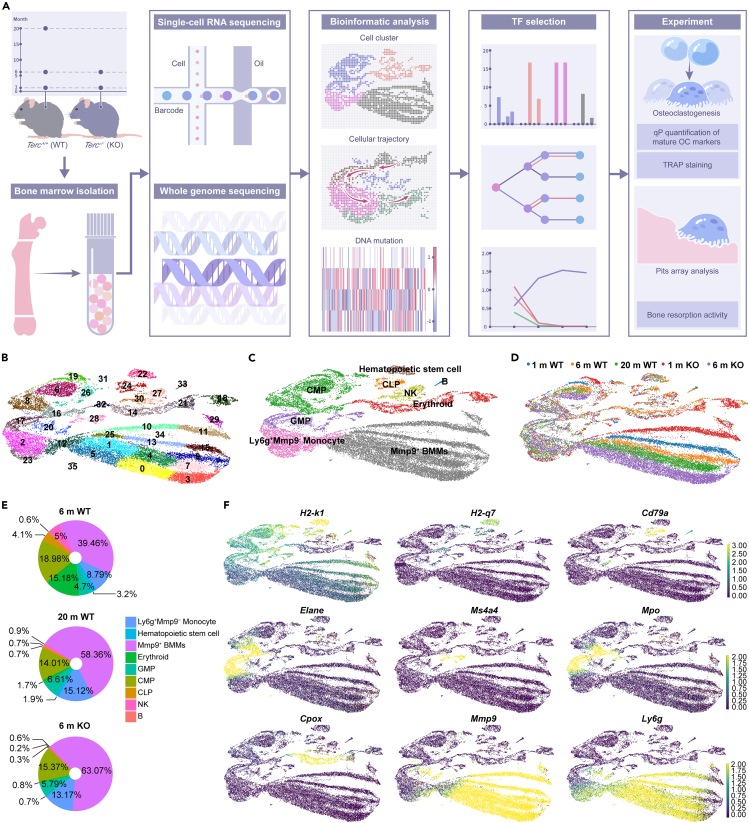
Figure 2.
Different osteoclast progenitor cell populations were identified based on the single-cell transcriptome analysis of bone marrow cells from aging wild-type and Terc−/− mice
See also Figure S2 and Table S6.
(A) The bone marrow cells from 1 m, 6 m, 20 m WT, and 1 m, 6 m Terc−/− mice were profiled with single-cell RNA-seq and DNA-seq. n = 8 (4 WT and 4 KO) for 1 m; n = 8 (4 WT and 4 KO) for 6 m; n = 4 (WT) for 20 m.
(B) is the UMAP plot that presents the cell clusters obtained from Leiden clustering.
(C) All the obtained clusters were annotated with the cell type information by comparing the top differential genes (signature genes) associated with each cluster with known cell type markers.
(D) Shows the cells from different libraries.
(E) Shows the cell compositions of 6 m WT, 20 m WT, and 6 m KO.
(F) List the UMAP plots for selected markers genes that we used for the cell type annotation.