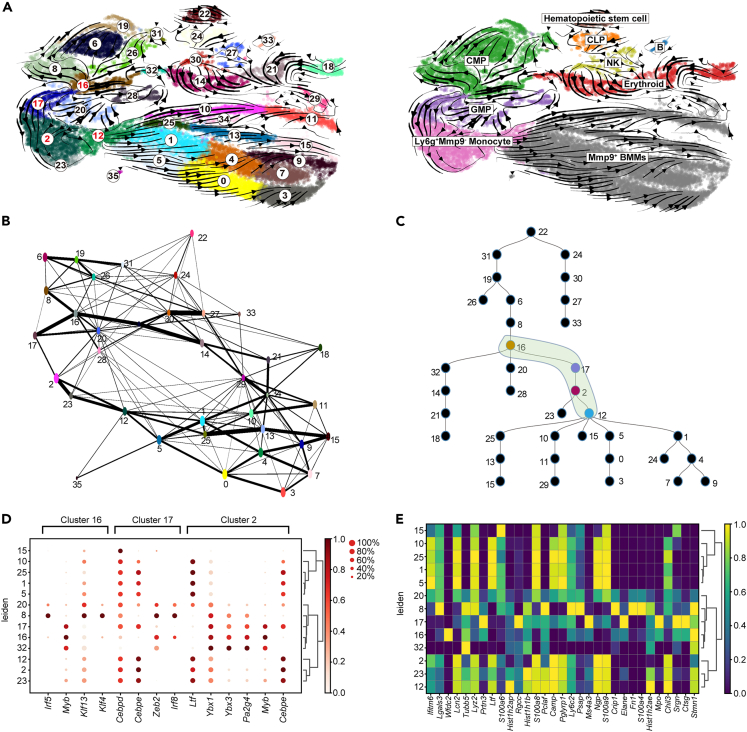
Figure 3.
The cellular trajectory reconstructed from mouse bone marrow aging single-cell RNA-seq data
(A) The RNA-velocity plot that we got from the single-cell RNA-seq data, which infers the cellular trajectory based on RNA velocity.
(B) A cellular trajectory (graph) inferred from the single-cell RNA-seq data by PAGA. This trajectory inference is based on the expression difference between cells, which is complementary to the RNA velocity based trajectory.
(C) Presents a cellular trajectory predicted by SCDIFF (version 2.0), which integrates the RNA velocity-based trajectory with the expression based trajectory (by PAGA). In the combined trajectory, we identified a key cellular transition stage for osteoclast cell differentiation (Cluster 16, 17, 2, and 12), which is marked in green in the plot.
(D) Shows the gene expression of top predicted transcription factors across different cell clusters of the osteoclast cellular trajectory.
(E) Shows the top signature genes associated with each of those clusters.