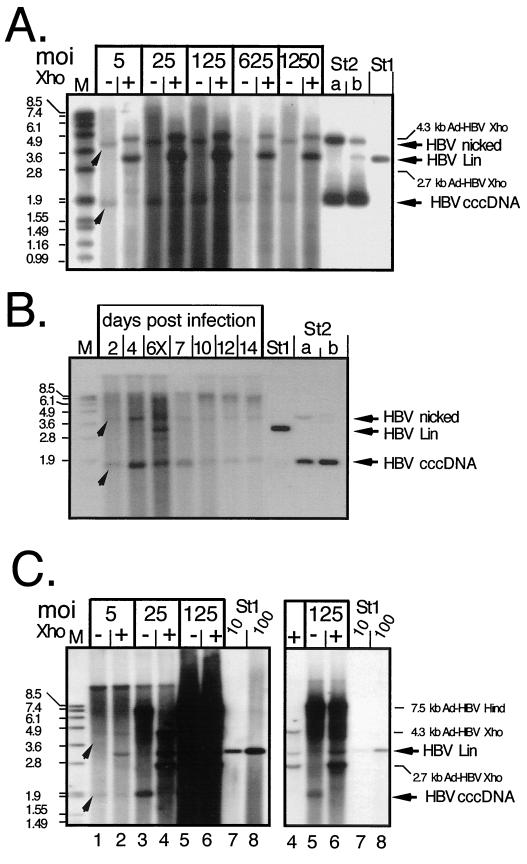
FIG. 7.
cccDNA generation in Ad-HBV-transduced PTHs and Huh7 cells. (A) Dose dependence in PTHs. Episomal DNAs were isolated at day 7 p.i. from PTHs transduced with Ad-HBV1.3 at the indicated MOIs and further treated to remove most noncircular molecules. One-half of each sample was directly loaded (lanes −), and the other half was loaded after incubation with XhoI (lanes +). HBV-specific DNAs were detected with a DIG-labeled probe. St1, 50 pg of linear 3.2-kb HBV DNA; St2, 3.2-kb pUC plasmid containing about 0.5 kb of HBV sequence. St2b contained 400 pg of directly loaded plasmid, and St2a contained 800 pg of plasmid that had been mixed with noninfected cell lysate and taken through the entire extraction procedure. The arrowheads indicate the positions of HBV cccDNA and its nicked form. As we found out later, the apparent signal increase after XhoI linearization was due to a specific cccDNA detection problem with the batch of agarose used in these experiments. (B) Time course in PTHs. PTHs were infected with Ad-HBV1.3 at an MOI of 100, and episomal DNAs were isolated at the indicated times. The sample from day 6 was partially digested with XhoI (lane 6X) to visualize the position of linear DNA. St1 and St2, reference DNAs as described for panel A. (C) Dose dependence in Huh7 cells. Episomal DNAs from cells infected at the indicated MOIs with Ad-HBV1.3 were analyzed as described above, except that during preparation HindIII rather than HpaI was used to cut DNAs not corresponding to HBV cccDNA. HindIII digestion of Ad-HBV1.3 generates a single HBV-containing fragment of 7.5 kb whose further cleavage by XhoI gives the same fragments as in panel A. The righthand gel shows a fivefold-shorter exposure of lanes 4 to 8, to resolve individual bands in the samples at an MOI of 125. St1, reference DNA (10 and 100 pg) as in panel A.