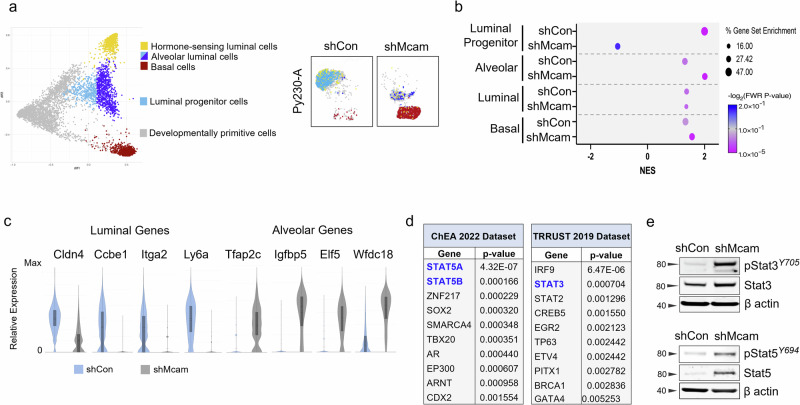
Fig. 3. A luminal progenitor to alveolar/basal lineage switch accompanies Mcam loss in Py230 cells.
a The top and bottom five differentially expressed genes between the adult mammary cell types identified in the diffusion map from Giraddi et al. (left)18, identify different cell states in Py230-A control and Mcam KD cells on the same scRNAseq coordinate map from Fig. 2c (right). b Gene set enrichment analysis (GSEA) showing enrichment of shCon and shMcam for LP and alveolar cell states, respectively compared to normal mouse mammary cell populations from Giraddi et al.18. c scRNA-seq analysis for relative expression distribution of luminal and alveolar cell state markers in Py230 control (blue) and Mcam KD (grey) cells. d Differentially expressed genes collectively overrepresented in shMcam-A compared to shCon-A Py230 cells analyzed for enrichment using the ChEA and TRRUST datasets from the Enrichr database. The top 10 enriched transcription factors expected to regulate shMcam upregulated genes in each analysis are shown. e Upregulated phosphorylation and expression of Stat3 and Stat5 in shMcam Py230-A cells relative to controls.