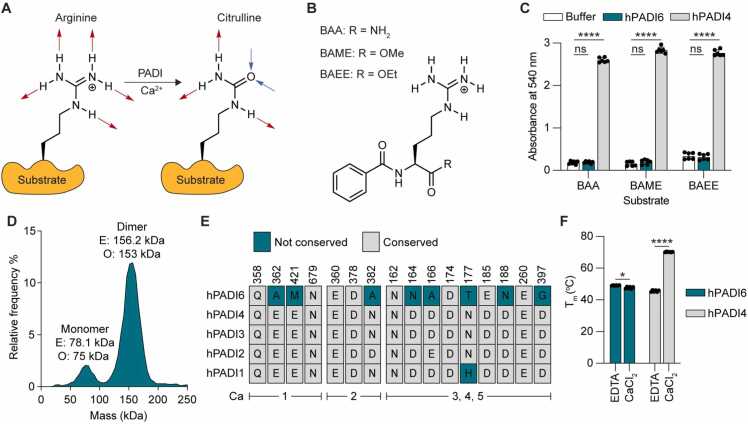
Fig. 1.
Biophysical and enzymatic characterisation of recombinant hPADI6. (A) Scheme of the PADI-catalysed post-translational conversion of the positively charged arginine to the neutral citrulline. Red arrows = hydrogen bond donors, blue arrows = hydrogen bond acceptors. (B) Structure of standard PADI substrates Nα-benzoyl-L-arginine amide (BAA), Nα-benzoyl-L-arginine methyl ester (BAME) and Nα-benzoyl-L-arginine ethyl ester (BAEE). (C) Activity of hPADI6 or hPADI4 (produced in-house from E. Coli[23]) with standard PADI substrates depicted in (B) measured using COLDER assays. Reactions performed in 10 mM CaCl2 and quenched after 1 h incubation at RT. [hPADI6] = 500 nM, [hPADI4] = 50 nM, [substrate] = 10 mM. Unpaired parametric t-test, * ** * = p < 0.0001. 2 independent replicates of 3 technical replicates performed. (D) Mass photometry histogram of hPADI6 expressed from Expi293 cells showing that hPADI6 mainly exists as a dimer in vitro. Bin size = 5 kDa. (E) Conservation of Ca2+ binding residues in human PADI enzymes, grouped by Ca site. Residue number in hPADI6 highlighted above. Not-conserved = teal, conserved = grey. (F) NanoDSF determined melting temperature (Tm) of hPADI6 and hPADI4 in either 10 mM EDTA or 10 mM CaCl2. Unpaired parametric t-test, * ** * = p < 0.0001. 2 independent replicates of 3 technical replicates performed.