Fig. 4.
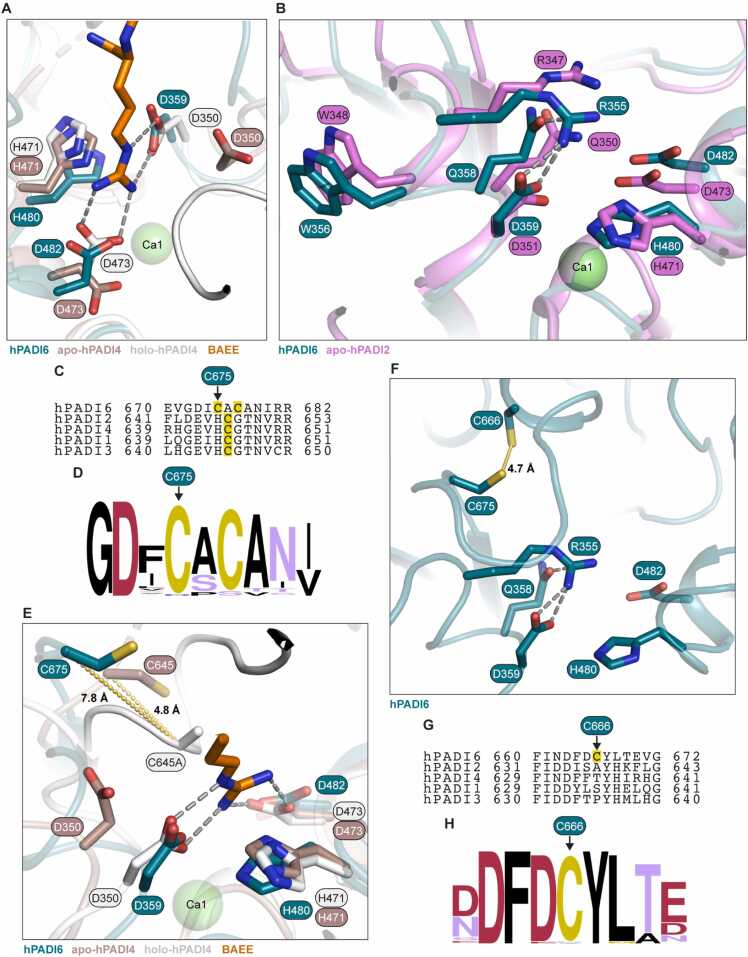
The hPADI6 active site partially aligns with hPADI2 and hPADI4. (A) Close up view of hPADI6 active site residues (teal), aligned with apo-hPADI4 (brown, PDB: 1WD8), and holo-hPADI4 (white, PDB: 1WDA). Substrate BAEE as part of the holo-hPADI4 displayed in orange. (B) Close up view of hPADI6 active site residues (teal), aligned with apo-hPADI2 (pink, PDB: 4N20). Key arginine, glutamine and tryptophan residues, as well as catalytic tetrad residues highlighted. (C) Sequence alignment of the human PADIs centred on the key catalytic cysteine residue. Yellow shading = potential hPADI6 key catalytic cysteine residues and confirmed hPADI1–4 catalytic cysteines. Predicted hPADI6 catalytic cysteine C675 highlighted. Sequence alignment produced using Clustal Omega [39]. (D) Logo plot of the hPADI6 sequence surrounding potential catalytic cysteine residues C675 and C677 aligned with the sequences of PADI6 in 79 other species. For a list of species used see S1 File. Logo plot produced using WebLogo.berkeley.edu [40], [41]. (E) Close up view of hPADI6 active site residues (teal), aligned with apo-hPADI4 (brown, PDB: 1WD8), and holo-hPADI4 (white, PDB: 1WDA). Predicted hPADI6 catalytic cysteine shown with distances to confirmed hPADI4 catalytic cysteine C645 (C645A in holo-hPADI4 structure) in the apo and holoenzyme structures. Substrate BAEE as part of the holo-hPADI4 structure displayed in orange. (F) Close-up view of hPADI6 C675 and C666 positioned above the catalytic aspartic acids and histidine. Distance between sulphur atoms of each cysteine = 4.7 Å. (G) Sequence alignment of the human PADIs centred on the hPADI6 C666, highlighted by yellow shading. C666 is not conserved in hPADIs 1 to 4. Sequence alignment produced using Clustal Omega [39]. (H) Logo plot of the PADI6 sequence surrounding hPADI6 C666 aligned with the PADI6 sequences of 79 other species. C666 is conserved in 78 out of 80 aligned sequences. For a list of species used see S1 File. Logo plot produced using WebLogo.berkeley.edu [40], [41].