Fig. 5.
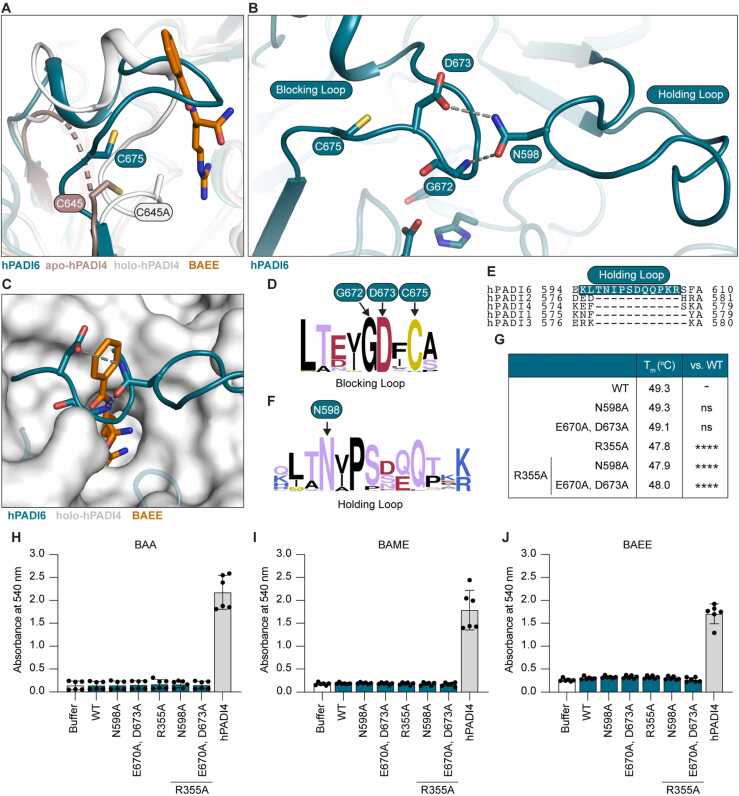
The hPADI6 active site pocket is blocked. (A) Close-up view of unstructured loop D632-C645 in hPADI4 in the absence of Ca2+ (brown, PDB: 1WD8) that becomes structured in holo-hPADI4 (white, PDB: 1WDA) aligned with corresponding loop in hPADI6 that is already structured in the absence of Ca2+ (teal). Unstructured loops displayed by dashed lines. Substrate BAEE as part of the holo-hPADI4 structure displayed in orange. hPADI4 catalytic cysteine C645 (C645A in holo-hPADI4 structure) and predicted hPADI6 catalytic C675 highlighted. (B) hPADI6 blocking and holding loops showing hydrogen bonds between D673 of the blocking loop and backbone NH of G672, with N598 of the holding loop. Predicted catalytic cysteine C675 is adjacent to the blocking loop. (C) Surface representation of the holo-hPADI4 active site cleft (white, PDB: 1WDA), superimposed with ribbon representation of hPADI6 (teal). Substrate BAEE as part of the holo-hPADI4 structure displayed in orange. hPADI6 D673, G672, and N598 shown with predicted hydrogen bonds represented by grey dashed lines. (D) Logo plot of the hPADI6 blocking loop sequence aligned with the sequences of PADI6 in 79 other species. For a list of species used see S1 File. D673 is conserved in all 80 of the aligned sequences. Logo plot produced using WebLogo.berkeley.edu [40], [41]. (E) Sequence alignment of the human PADIs covering the hPADI6 holding loop sequence showing it is an insertion in the PADI6 sequence. hPADI6 holding loop highlighted with grey dashed box. Sequence alignment produced using Clustal Omega [39]. (F) Logo plot of the hPADI6 holding loop sequence aligned with the sequences of PADI6 in 79 other species. For a list of species used see S1 File. N598 is conserved in 79 of the 80 aligned sequences. Logo plot produced using WebLogo.berkeley.edu [40], [41]. (G) NanoDSF determined melting temperatures (Tm) of wild-type (WT) hPADI6 and variants. Unpaired parametric t-test, **** = p < 0.0001. 2 independent replicates of 3 technical replicates performed. (H) Activity of WT hPADI6, hPADI6 variants or hPADI4 with PADI substrate BAA determined by COLDER assay. Reactions performed in 10 mM CaCl2 and quenched after 1 h incubation at RT. [hPADI6/hPADI6 variant] = 500 nM, [hPADI4] = 50 nM, [substrate] = 10 mM. Unpaired parametric t-test, **** = p < 0.0001. 2 independent replicates of 3 technical replicates performed. (I) As (H) with BAME instead of BAA. (J) As (H) with BAEE instead of BAA.