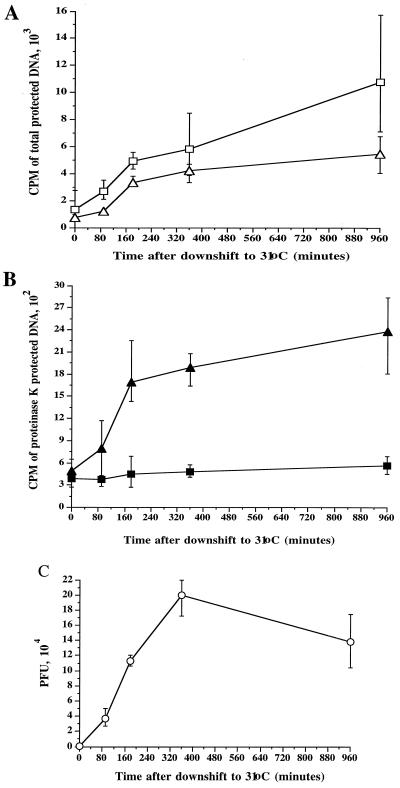
FIG. 1.
Measuring DNA packaging and capsid envelopment using a TCA precipitation assay. HuH7 cells were infected with the HSV strain tsProt.A, acid washed, and then incubated at 39°C in the presence of [3H]thymidine. After 6.5 h, cycloheximide was added, and after a further 30 min the cells were downshifted to 31°C. Total cell extracts were prepared at particular times after downshift (indicated on the horizontal axes) and measured as follows. (A) DNase I-resistant (total packaged) DNA in the presence (squares) or absence (triangles) of 0.05% Triton X-100. (B) Proteinase K/DNase I-resistant (enveloped) DNA in the presence (solid squares) or absence (solid triangles) of 0.05% Triton X-100. (C) PFU present in each extract. In each case, plotted values represent the mean of two to four samples, and error bars indicate the range from the mean.