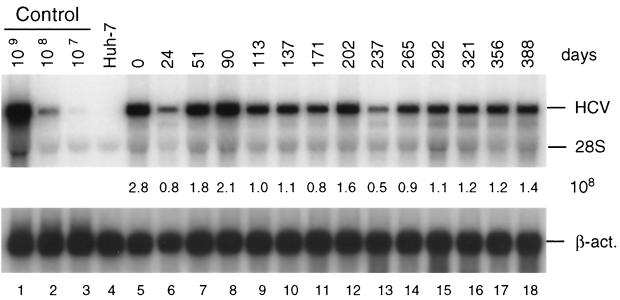
FIG. 2.
Stability of HCV replicon in cell line 9-13 passaged under continuous G418 selection. Cells were regularly passaged three times a week in the presence of G418 (1 mg/ml) and total RNA was isolated from cells at given time points. After denaturing agarosegel electrophoresis, HCV- and β-actin-specific RNAs were detected by Northern bloting using 32P-labeled riboprobes complementary to the HCV IRES and neo or a β-actin-specific antisense RNA, respectively, and quantified by phosphorimaging. The number of replicon RNA molecules was determined by comparison with the serial dilution of in vitro transcripts (lanes 1 to 3). β-Actin RNA served as a control to correct for the amount of total RNA loaded in each lane of the gel (∼2 μg). The result obtained with total RNA from the parental Huh-7 cells is shown in lane 4. Numbers between both panels refer to the number of replicon RNA molecules (108) contained in 1 μg of total RNA of the respective sample. Note that the control RNA is a mixture of given numbers of in vitro transcripts and 2 μg of total RNA from the parental Huh-7 cells that was used as carrier. The positions of HCV RNA, 28S rRNA, and β-actin mRNA (β-act.) are given to the right. Analogous results were obtained with cell line 5-15 (not shown).