Extended Data Fig. 10: Comparing 1-shot similarity heatmaps of pretrained encoders with class prototype.
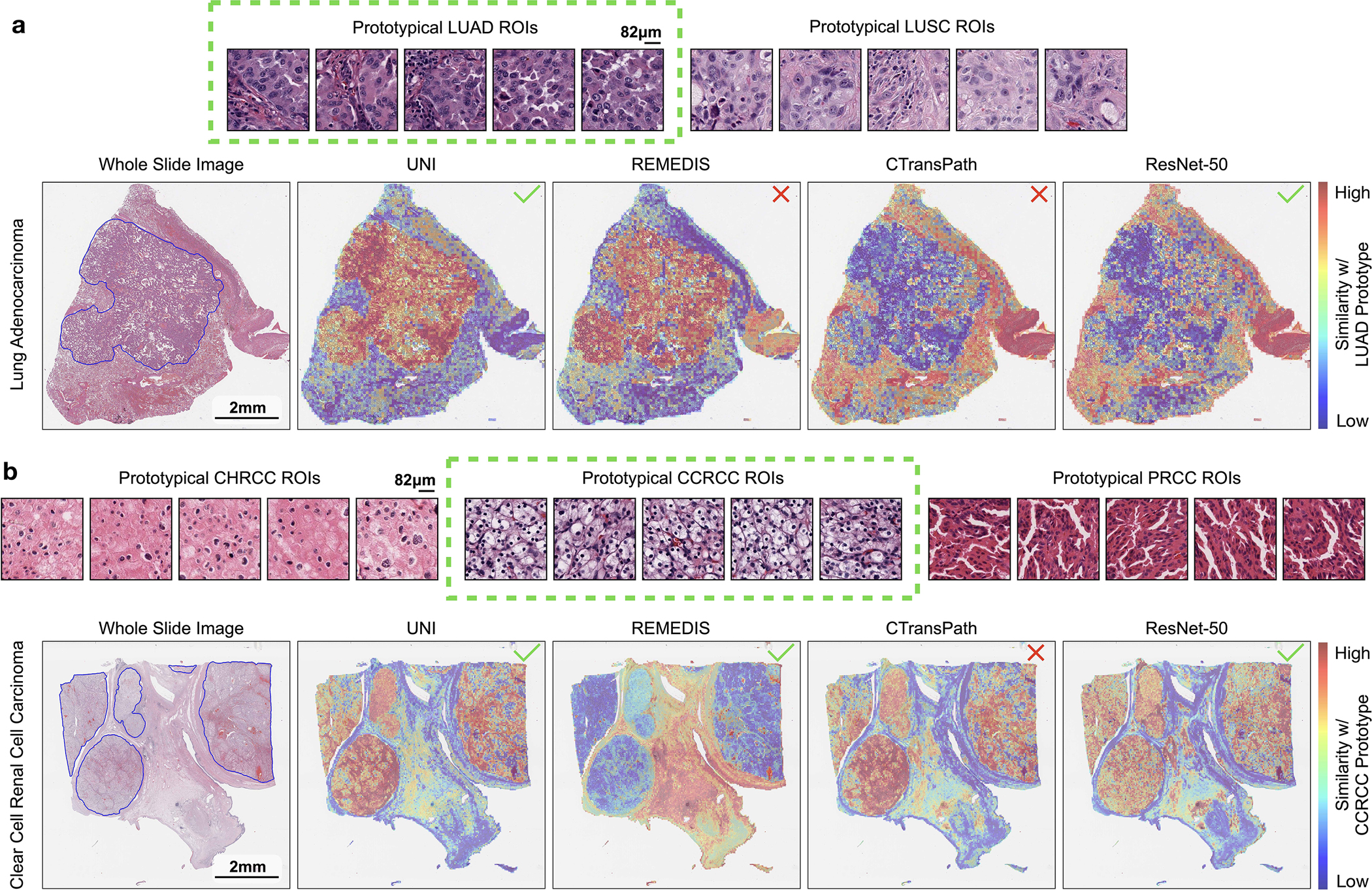
We compare the similarity heatmaps of all pretrained encoders using annotated ROIs from a single slide per class for forming class prototypes in MI-SimpleShot (with top-5 pooling) on NSCLC subtyping (a) and RCC subtyping task (b), with top visualizing example ROIs used for each class, and bottom showing similarity heatmaps. Outlined in blue are pathologist annotations of ROIs that match the label of the histology slide. Similarity heatmaps are created with respect to the class prototype of the correct slide label (indicated in green), with a ✓ indicating a correct prediction and ✗ indicating incorrect prediction. Note that since the visualizations are created with respect to the ground truth label, the model may retrieve correct patches that match pathologist annotations but still misclassify the slide. a. On a LUAD slide, we observe strong agreement of the pathologist’s annotations with retrieved LUAD patches by UNI. Although retrieved patches by REMEDIS also have strong agreement with the pathologist’s annotations, we note that slide was misclassified as LUSC, indicating that the top-5 retrieved patches of the LUSC prototype was higher than that of the LUAD prototype. Vice versa, ResNet-50IN classifies the slide correctly but incorrectly retrieves the correct patches that agree with the pathologist’s annotations, indicating that non-LUAD patches in the slide were more LUAD-like than the pathologist-annotated LUAD patches with respect to the LUAD prototype. The similarity heatmap for CTransPath both misclassified the slide and retried incorrect patches. b. On an CCRCC slide, we observe strong agreement of the pathologist’s annotations with retrieved CCRCC patches by UNI. We observe similar mismatch in predicted class label and retrieved patches, in which REMEDIS classifies the slide correctly but retrieves the incorrect patches, and CTransPath misclassifies the slide but retrieves the correct patches.
