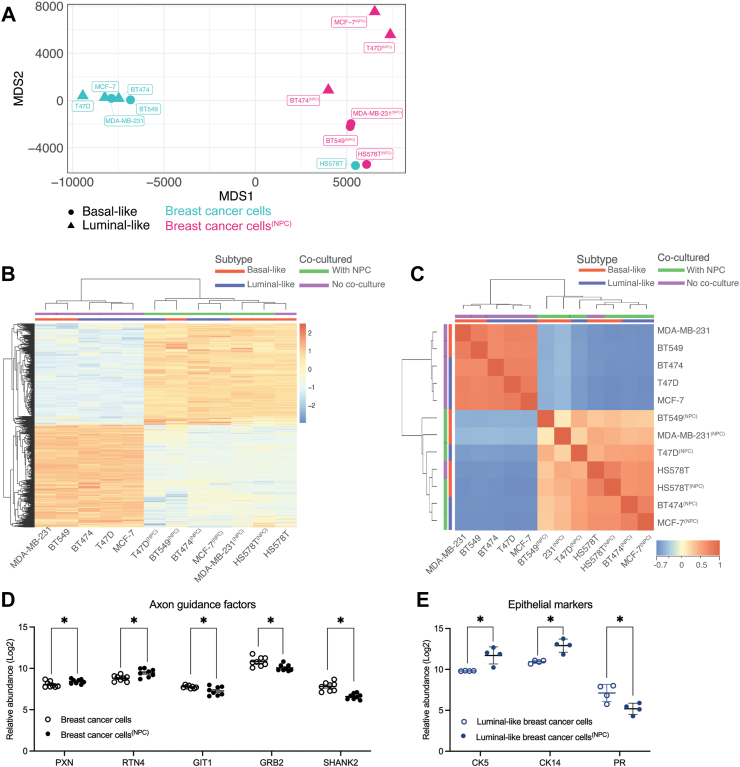
Fig. 4.
Proteomic landscape in breast cancer cells before and after NPC co-culture. a Multidimensional scaling plot displaying the breast cancer cell proteome after the selection of statistically significant proteins before and after co-culture with NPCs (Benjamini Hochberg (BH) adjusted FDR < 0.05), n = 6 breast cancer cell lines. Superscripted text indicates the co-culture conditions. b Hierarchical clustering of normalized TMT-ratios pooling breast cancer cells together, except for HS578T, when comparing statistically significant proteins before and after co-culture with NPCs (BH adjusted FDR < 0.05). Colour bar: Blue represents a decreased protein expression compared to the mean intensity with a gradient into beige for no change in expression and into red for proteins with increased expression. c Correlation plot of breast cancer cells comparing statistically significant proteins before and after co-culture with NPCs (BH adjusted FDR < 0.05). Each square represents the Spearman correlation coefficient between two samples with comparisons hierarchically clustered. Colour bar: Blue represents a negative correlation with a gradient into beige for no correlation and into red for positive correlation. d Selected proteins involved in axon guidance with changed expression pattern before and after co-culture with NPCs (∗p ≤ 0.05, significance was determined by unpaired t-test). e Expression of epithelial markers - cytokeratin (CK14, CK5) and progesterone receptor (PR) in luminal-like breast cancer comparing before and after co-culture with NPCs (∗p ≤ 0.05, significance was determined by unpaired t-test).