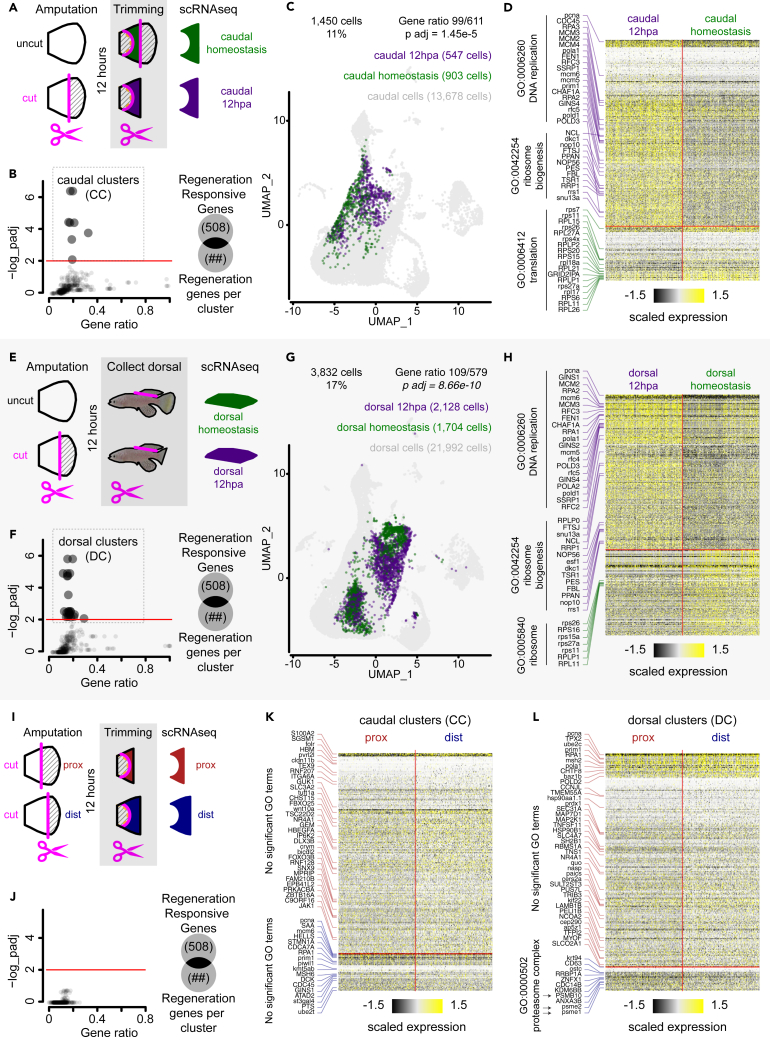
Figure 3.
Amputation induces regeneration transcriptional response within epidermis subpopulation at 12 hpa
(A) Experimental design of 12 hpa and homeostasis tissue collection for scRNA-seq on the caudal fin. Fins were uncut or 50% cut, 12 h later the stump was trimmed to match the corresponding anatomical region between the two samples, then tissues were dissociated and subjected to scRNA-seq.
(B) RRG enrichment analysis on DE genes between 12 hpa and homeostasis caudal cells split by clusters defined in the integrated dataset (656 clusters); Fisher’s exact test, FDR < 0.01.
(C) UMAP distribution of caudal clusters (CCs) within the caudal dataset, cell numbers per sample, and RRG enrichment analysis; Fisher’s exact test, p < 0.001.
(D) Heatmap of DE genes between caudal 12 hpa and caudal homeostasis, and top GO terms associated with the gene list (FDR < 0.05). Cells were randomly sampled to balance both 12 hpa and homeostasis groups.
(E) Experimental design of 12 hpa and homeostasis intact dorsal fin collection for scRNA-seq. Caudal fins were uncut or 50% cut, 12 h later the intact dorsal fin was collected, dissociated, and subjected to scRNA-seq.
(F) RRG enrichment analysis on DE genes between 12 hpa and homeostasis dorsal cells split by clusters defined in the integrated dataset (656 clusters); Fisher’s exact test, FDR < 0.01.
(G) UMAP distribution of dorsal clusters (DC) within the dorsal dataset, cell numbers per sample, and RRG enrichment analysis; Fisher’s exact test, p < 0.001.
(H) Heatmap of DE genes between dorsal 12 hpa and dorsal homeostasis, and top GO terms associated with the gene list (FDR < 0.05). Cells were randomly sampled to balance both 12 hpa and homeostasis groups.
(I) Experimental design of proximal and distal regenerating caudal fins collected for scRNA-seq. Fins were amputated either proximal or distal, 12 h later the stump was trimmed to remove the scales and muscle, then tissues were dissociated and subjected to scRNA-seq.
(J) RRG enrichment analysis on DE genes between proximal and distal cells split by clusters defined in the integrated dataset (656 clusters); Fisher’s exact test, FDR < 0.01.
(K) Heatmap of DE expressed genes between proximal and distal cells within the caudal clusters (CCs). No significant GO terms were found (FDR < 0.05). Cells were randomly sampled to balance both proximal and distal groups.
(L) Heatmap of DE expressed genes between proximal and distal cells within the dorsal clusters (DC), and single GO terms associated with the gene list (FDR < 0.05). Cells were randomly sampled to balance both proximal and distal groups.