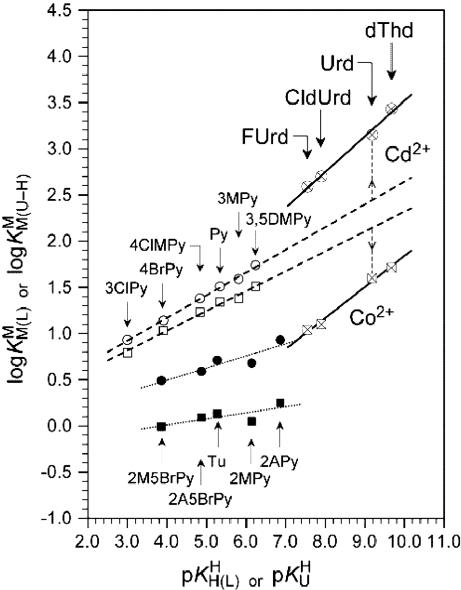
Fig. 2.
Comparison of the 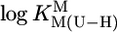 vs.
vs. 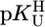 relationship (⊗, ⊠) for the uridinate-type (U–H)– ligands shown in Fig. 1 for their Co2+ (⊠, □, ▪) and Cd2+ (⊗, ○, •) complexes with the corresponding
relationship (⊗, ⊠) for the uridinate-type (U–H)– ligands shown in Fig. 1 for their Co2+ (⊠, □, ▪) and Cd2+ (⊗, ○, •) complexes with the corresponding 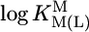 vs.
vs. 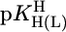 relationships for PyN [L = PyN = 3-chloropyridine (3ClPy), 4-bromopyridine (4BrPy), 4-(chloromethyl)pyridine (4ClMPy), pyridine (Py), 3-methylpyridine (3MPy), and 3,5-dimethylpyridine (3,5DMPy) (from left to right)] (○, □) and for o-substituted PyN [L = oPyN = 2-methyl-5-bromopyridine (2M5BrPy), 2-amino-5-bromopyridine (2A5BrPy), tubercidin (7-deazaadenosine, Tu), 2-methylpyridine (2MPy) and 2-aminopyridine (2APy)] (•, ▪). The reduced stability of the M(oPyN)2+ complexes reflects the steric inhibition of an o-amino or o-methyl group. The data pairs for the M(U–H)+ complexes are from Table 1, and those for the M(PyN)2+ and M(oPyN)2+ species are from table 3 in ref. 27. The least-squares straight reference lines are drawn according to Eq. 5 (see Table 3 and ref. 27). The plotted equilibrium constants (aq solution; 25°C) for the M2+/(U–H)– systems (Table 1) refer to I = 0.1 M (NaNO3), and those for the M2+/PyN or oPyN systems refer to I = 0.5 M (NaNO3); this change in I from 0.1 to 0.5 M is of no significance, because the latter ligands do not carry a charge, and the shifts in
relationships for PyN [L = PyN = 3-chloropyridine (3ClPy), 4-bromopyridine (4BrPy), 4-(chloromethyl)pyridine (4ClMPy), pyridine (Py), 3-methylpyridine (3MPy), and 3,5-dimethylpyridine (3,5DMPy) (from left to right)] (○, □) and for o-substituted PyN [L = oPyN = 2-methyl-5-bromopyridine (2M5BrPy), 2-amino-5-bromopyridine (2A5BrPy), tubercidin (7-deazaadenosine, Tu), 2-methylpyridine (2MPy) and 2-aminopyridine (2APy)] (•, ▪). The reduced stability of the M(oPyN)2+ complexes reflects the steric inhibition of an o-amino or o-methyl group. The data pairs for the M(U–H)+ complexes are from Table 1, and those for the M(PyN)2+ and M(oPyN)2+ species are from table 3 in ref. 27. The least-squares straight reference lines are drawn according to Eq. 5 (see Table 3 and ref. 27). The plotted equilibrium constants (aq solution; 25°C) for the M2+/(U–H)– systems (Table 1) refer to I = 0.1 M (NaNO3), and those for the M2+/PyN or oPyN systems refer to I = 0.5 M (NaNO3); this change in I from 0.1 to 0.5 M is of no significance, because the latter ligands do not carry a charge, and the shifts in 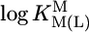 and
and 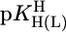 go “parallel” to each other (27).
go “parallel” to each other (27).