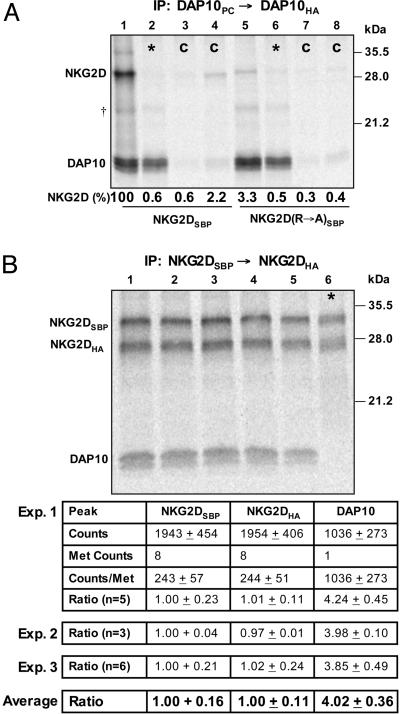
Fig. 1.
Stoichiometry of the NKG2D–DAP10 complex. (A) An in vitro translation system with ER microsomes was used to study the assembly of NKG2D and DAP10. After assembly, membranes were solubilized with 0.5% digitonin, and 35S-labeled complexes were isolated in a two-step snIP that targeted the PC and HA epitope tags attached to the two DAP10 chains. In the mixing controls (*, lanes 2 and 6), DAP10 and NKG2D chains were translated separately, and the reactions were combined immediately before solubilization. In the antibody controls (C, lanes 3, 4, 7, and 8), the PC antibody (lanes 3 and 7) or the HA antibody (lanes 4 and 8) used in the first and second IP step, respectively, were replaced with species- and isotype-matched antibodies of irrelevant specificity. Assembly was abrogated by mutation of the TM arginine of NKG2D to alanine (NKG2D R→ A, lane 5). The ratio of NKG2D and DAP10 dimer was determined and expressed as the percentage relative to WT (lane 1). Data are representative of four experiments. †, A faint background band appeared at the indicated position in this and other experiments; this band was nonspecific because it was also observed in mixing controls. (B) The stoichiometry of the complex was directly determined by quantification of 35S-labeled NKG2D and DAP10 with a phosphorimager. The NKG2D dimer was isolated with a two-step snIP that targeted the SBP and HA tags attached to the two NKG2D chains, and the molar ratio between NKG2D and associated DAP10 was determined based on the known methionine content of the chains. Multiple parallel reactions were analyzed in each of the three experiments, and the average and standard deviation were calculated for all reactions. *, In each experiment, a mixing control was included to verify specificity. These experiments demonstrated that four DAP10 chains are present for each NKG2D dimer.