Fig. 2.
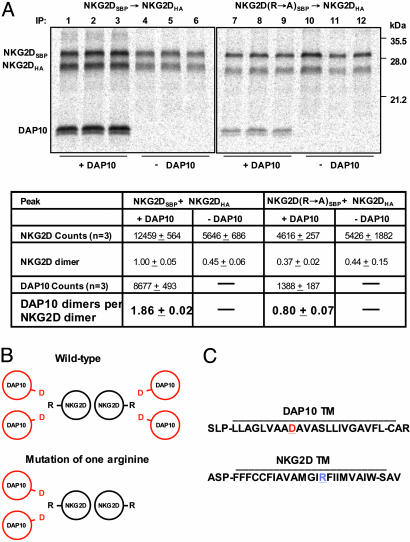
Cooperative assembly of the hexameric NKG2D–DAP10 complex. (A) NKG2D dimers composed of two WT chains (lanes 1–6) or of one WT and one mutant chain (substitution of TM arginine by alanine, R→ A) (lanes 7–12) were isolated by a two-step snIP that targeted the SBP and HA tags attached to the two NKG2D chains. The DAP10 construct used for this experiment had two additional methionine residues (total of three methionine residues, compared with one for WT DAP10 used in Fig. 1) to increase the signal. The stoichiometry measurements are presented here for NKG2D dimers and DAP10 dimers, rather than individual chains as in Fig. 1. Mutation of one of the TM arginine residues in the NKG2D dimer reduced the number of associated DAP10 dimers from 1.86 for the WT NKG2D dimer (lanes 1–3) to 0.8 (lanes 7–9), indicating that only a single DAP10 dimer was bound to the mixed NKG2D dimer. Furthermore, the yield of the mixed NKG2D dimer was reduced to 37% (lanes 7–9) relative to WT (100%, lanes 1–3). In reactions without DAP10 (lanes 4–6 and 10–12), the yield of NKG2D dimer was reduced to a level similar to the NKG2D dimer with one mutated chain. Data are representative of three experiments. (B) Graphical representation of the TM domains (as simplified helical wheels) for the six-chain complex formed by WT NKG2D dimer, compared with the four-chain complex formed by the mixed NKG2D dimer in which one of the TM arginine residues (R) has been mutated to alanine (A). The aspartic acid (D) residues in the TM domains of the DAP10 dimer are indicated in red and positioned at the interface with the NKG2D TM domains. (C) Predicted TM sequences and flanking segments for NKG2D and DAP10.