Fig. 3. Tailocins target closely related pathogens.
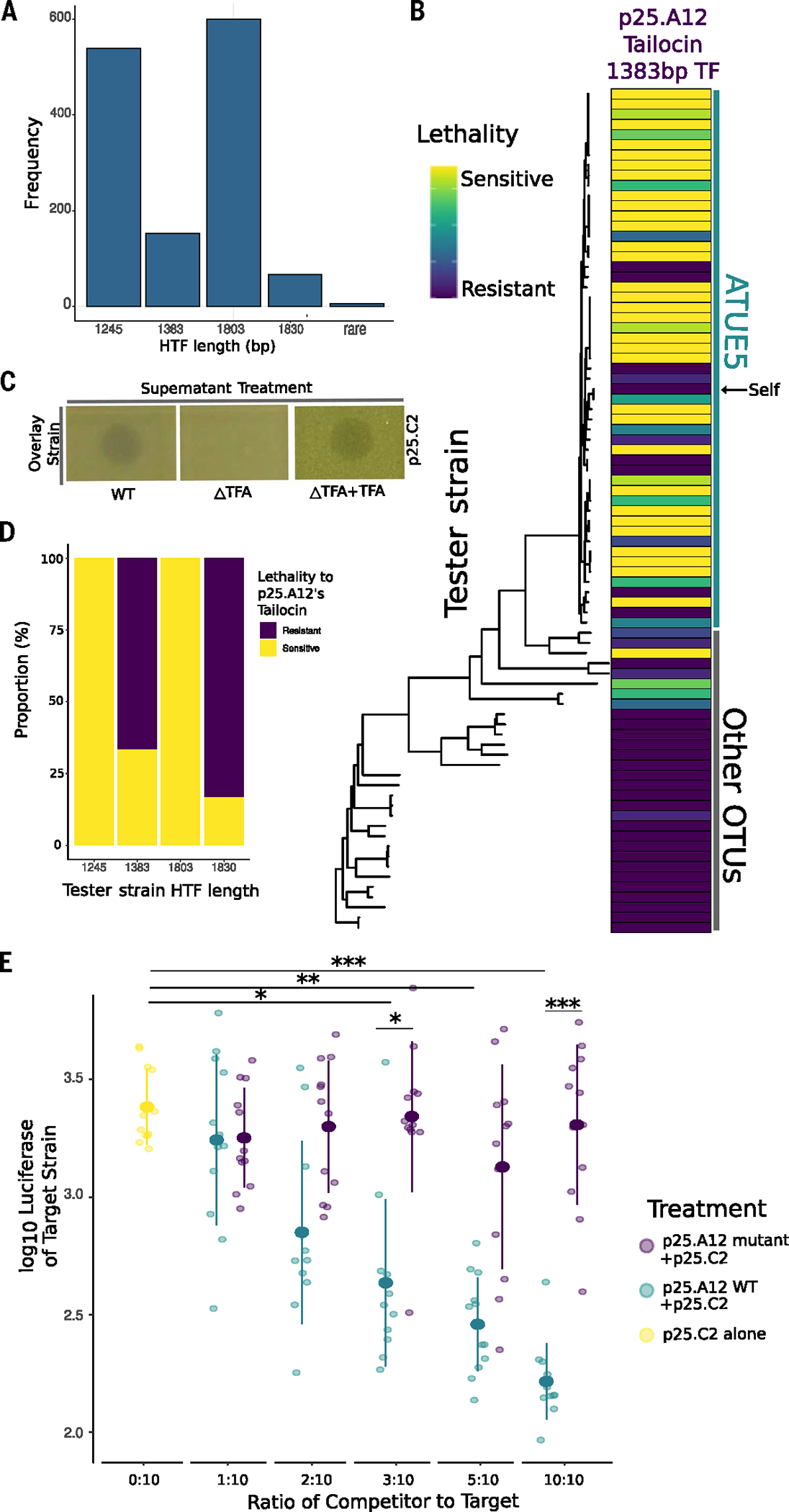
(A) Frequency of HTF nucleotide sequence lengths. There are four highly conserved lengths within the Pseudomonas populations. (B) Tailocins are preferentially used for intralineage killing. Soft agar cultures of the Pseudomonas strains (rows) were challenged with viral particles extracted from cultures of one strain, p25.A12 (from the 1383-bp hypothetical tail fiber length haplotype, column), in three technical and three biological replicates. The phylogeny includes 83 Pseudomonas representative strains and is displayed according to phylogenetic placement. Vertical lines indicate strains that belong to ATUE5 (green) and other OTUs (gray). Interactions with the strain’s own tailocin are indicated by the black arrow pointing to self. For each replicate, a strain was given a score of 3 for clear zone of inhibition, a 2 for semiclear, a 1 for opaque, or a 0 for no killing, and then added together after all three replicates. (C) Knockout of the tail fiber assembly gene disrupts tailocin bactericidal activity. Killing activity is indicated by a clearing in the lawn of the overlay strain. Complementation of the gene on an overexpression plasmid in the knockout strain restores the killing phenotype. (D) The proportion of tester strains sensitive or resistant to p25.A12’s tailocin, a 1383-bp hypothetical tail fiber, significantly correlated with the tester strain’s hypothetical tail fiber length haplotype (Fisher’s exact test, P = 10–8). (E) In planta coinfections of p25.A12 (competitor) and p25.C2 (target, known to be sensitive to p25. A12). Different ratios of competitor and constant amounts of target strain were used. p25.C2 was grown alone as the control. ANOVA test P values are shown. ***P < 0.001, **P < 0.01, or *P < 0.05.
