Fig. 5. The same length haplotypes of HTF are present in contemporary and century-old historical samples, and the HTF haplotype diversity is maintained at the leaf scale and broader geographical scales.
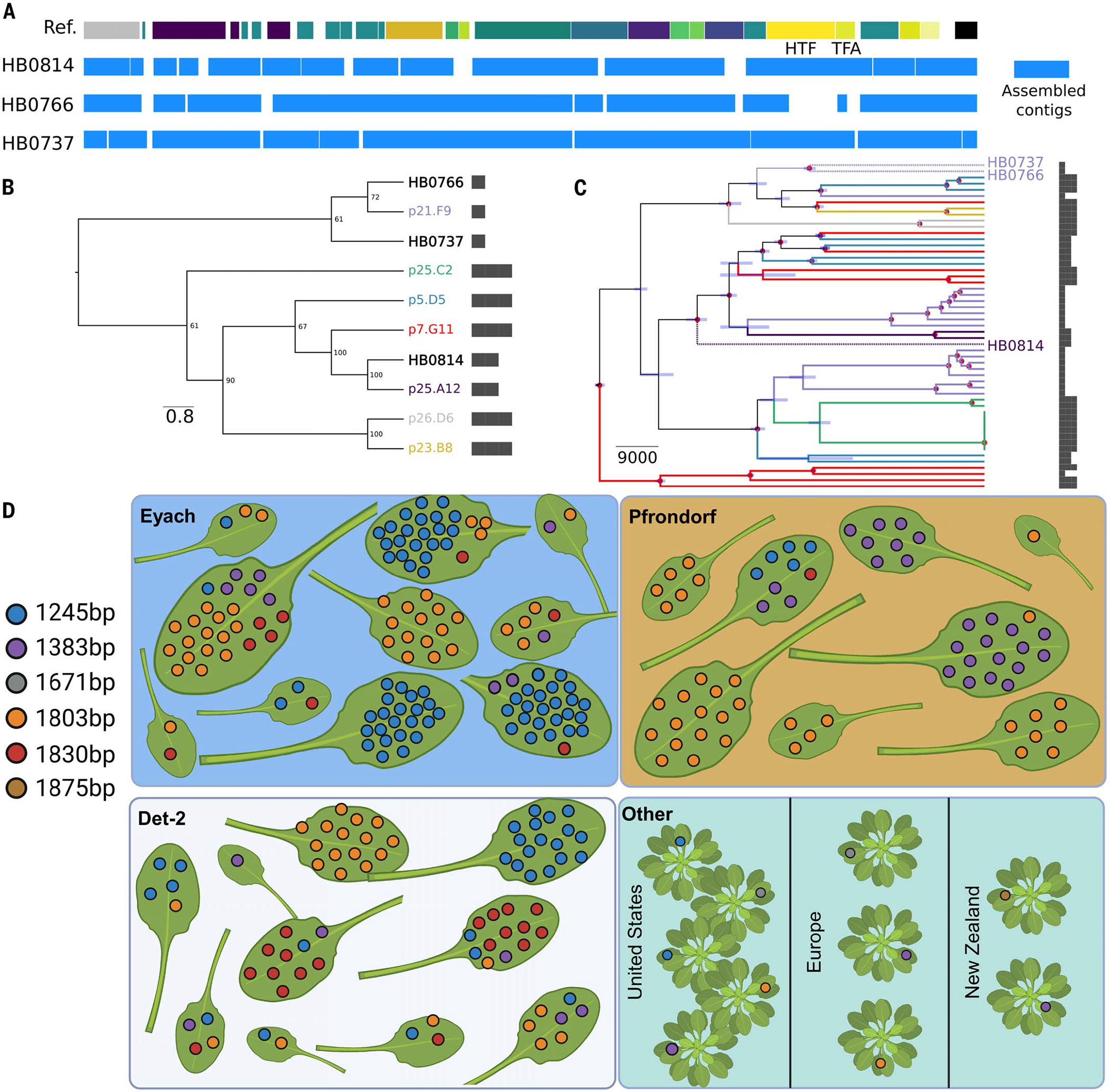
(A) De novo assembled contigs spanning the tailocin genomic region for the historical samples. The reference track on the top represents the genes present in the ATUE5 reference genome. The blue boxes depict the de novo assembled contigs. (B) Maximum-likelihood neighbor-joining tree of the HTF gene translated sequence. Historical samples (HB) are placed in the context of the most common haplotypes. HTF lengths are shown with bars. (C) Bayesian tip-date calibrated phylogeny representing the evolutionary relationship between historical and modern Pseudomonas strains. The tips and branches are colored based on the HTF gene haplotype. Only historical sample labels are shown with stars. The node bars represent the 95% highest posterior density intervals of the estimated time, and the nodes marked with red dots represent those with a posterior probability of 1. HTF lengths are shown with bars. (D) A subset of eight to 10 plants from the three German populations (blue, orange, and white panels) and 10 P. syringae isolates collected from plants globally, which were downloaded from NCBI (green). Each leaf is from a single plant. Colored dots indicate a single bacterial isolate found and its corresponding HTF length. Plants in the bottom right panel indicate those not found within the same population. [Figure created with BioRender.com]
