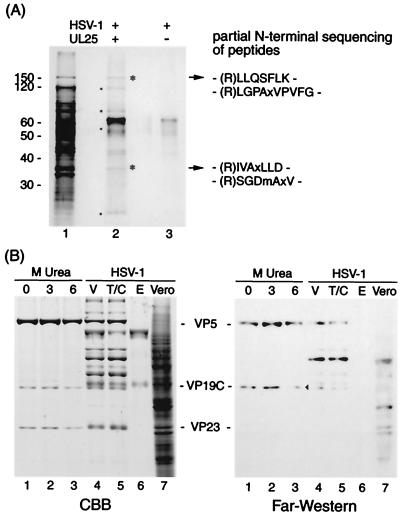
FIG. 3.
Protein-protein interaction of UL25 with virus proteins. (A) Identification of virus proteins reconstituted with UL25 on sheets; (B) far-Western analysis of UL25 binding to virus proteins. (A) A mixture of virus proteins solubilized with 8.0 M urea and PVDF sheets blotted with UL25 (lane 2) or not (lane 3) were renatured by stepwise dialysis. This dialysis allows virus proteins to bind UL25 protein on PVDF by reconstitution of interactive proteins. Virion proteins (lane 1) and proteins reconstituted on UL25 sheets (lane 2) were analyzed by SDS-PAGE. Asterisks and dots indicate the proteins reconstituted with UL25. Following trypsin digestion of bound proteins (asterisks in lane 2), the result obtained by amino-terminal sequencing of the digested peptides is shown on the right. The 65-kDa band appears to be more intense in lane 2 than in lane 3, since bands of UL25 and BSA are overlapping. The migration of molecular mass markers is shown on the left. (B) HSV-1 B capsids were extracted with 0 (lane 1), 3.0 (lane 2), and 6.0 (lane 3) M urea. These extracts (lane 1 to 3), Vero cells (lane 7), purified virions (V, lane 4), and their tegument-capsid (T/C, lane 5) and envelope (E, lane 6) components were subjected to SDS-PAGE (CBB stain) and far-Western analysis using biotin-labeled UL25. An arrowhead indicates the VP19C band.