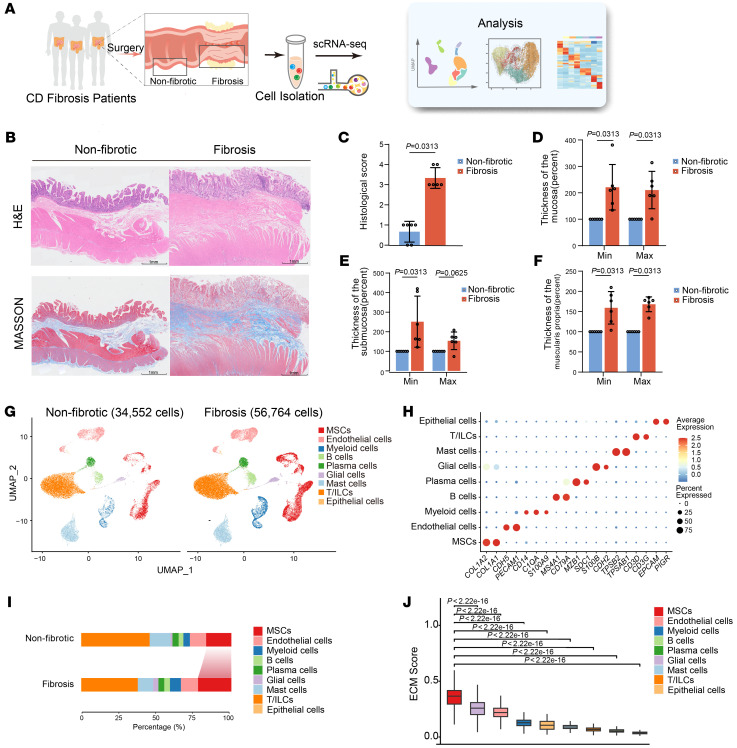
Figure 1. Cellular landscape of fibrotic and nonfibrotic tissues from patients with intestinal fibrosis.
(A) Graphic overview of the study design. Surgical specimens of fibrotic and adjacent nonfibrotic intestinal segments from patients with CD were processed into single-cell suspensions and subjected to scRNA-Seq using 10x Genomics. Integrated analyses of single-cell transcriptome data are shown in the rectangle to the right. (B) Representative plots of H&E and Masson’s trichrome staining of fibrotic and nonfibrotic intestine tissues from a patient with CD. Scale bar: 1 mm. (C) Bar plots showing histologic scores of the fibrotic intestinal (n = 6) and nonfibrotic (n = 6) segments. Data represent the mean ± SD. Statistical differences were determined by paired Wilcoxon’s rank-sum tests. (D–F) The relative minimal (min) and maximal (max) width of the mucosa (D), submucosa (E), and muscularis propria (F) of the fibrotic and nonfibrotic intestine. All nonfibrotic values were normalized to 100% to calculate the relative thickness of the fibrosis site. Data represent the mean ± SD. Statistical differences were determined by paired Wilcoxon’s rank-sum tests. (G) Uniform manifold approximation and projection (UMAP) plots showing 9 major cell types from 6 fibrotic samples (56,764 cells) and 6 nonfibrotic samples (34,552 cells). (H) Dot plots of representative markers in the indicated major cell types. The average gene expression and percentage of cells expressed are shown by dot color and size, respectively. (I) Bar graph showing the percentage of major cell types in fibrotic and nonfibrotic samples. (J) Box plots showing the ECM signature score of each cell type in fibrotic states. Statistical differences were determined by 1-way ANOVA with Bonferroni’s correction.