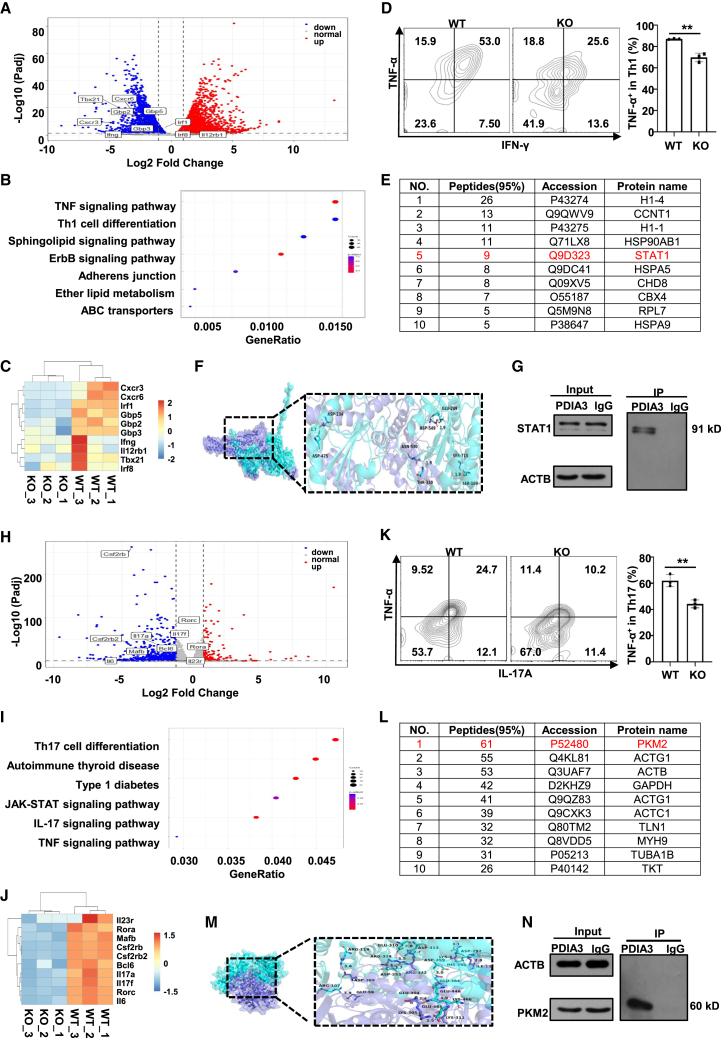
Figure 6.
PDIA3 interacts with STAT1 or PKM2 in effector T cells
(A–C) RNA-seq revealed differentially expressed genes (DEGs) in WT vs. KO Th1 cells. (A) Volcano plot showing DEGs between WT and KO Th1 cells. (B) KEGG pathway analysis on DEGs between WT and KO Th1 cells. (C) Representative heatmap results for selected DEGs between WT and KO Th1 cells. (D) Representative flow cytometry staining of INF-γ and TNF-α in differentiated Th1 cells from WT and KO mice. (E) Top 10 candidate binding partners of PDIA3 in Th1 cells. (F) Representative image of autodocking between PDIA3 and STAT1 protein. (G) Immunoprecipitation assay of the lysates of WT Th1 cells using anti-PDIA3 antibody. (H–J) RNA-seq revealed differentially expressed genes (DEGs) in WT vs. KO Th17 cells. (H) Volcano plot showing DEGs between WT vs. KO Th17 cells. (I) KEGG pathway analysis on DEGs between WT vs. KO Th17 cells. (J) Representative heatmap results for selected DEGs between WT vs. KO Th17 cells. (K) Representative flow cytometry results of IL-17A and TNF-α in differentiated Th17 cells from WT and KO mice. (L) Top 10 candidate binding partners of PDIA3 in Th17 cells. (M) Representative image of autodocking between PDIA3 and PKM2 protein. (N) Immunoprecipitation assay of the lysates of WT Th17 cells using anti-PDIA3 antibody. All in vitro studies were repeated at least three times. Data are expressed as mean ± SEM. Statistical significance was analyzed by unpaired Student’s t test. ∗∗p < 0.01.