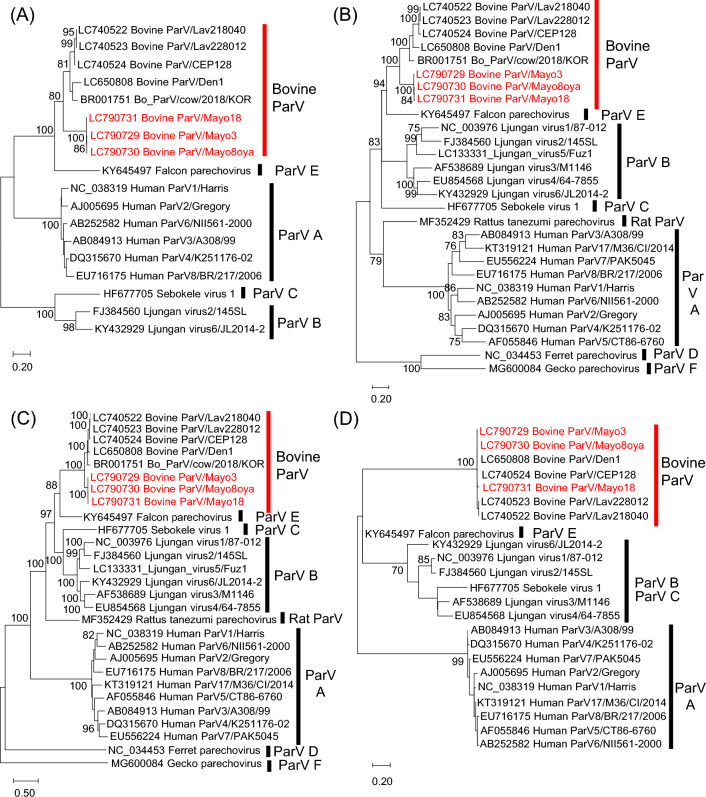
Fig. 1.
Phylogenetic analysis based on nucleotide sequences of the 5′UTR (A), VP1 region (B), 2C+3CD region (C), and 3′UTR (D) of the Bo_ParVs identified in this study (red text) and of other parechoviruses, obtained from the DDBJ/EMBL/GenBank database. Phylogenetic trees were constructed using the maximum-likelihood method in MEGA7 with best-fit models (K2+G for the 5′UTR, T92+G+I for VP1, GTR+G for 2B+3CD, and HKY for the 3′UTR). Bootstrap values above 70 (1000 replicates) are shown. The bars represent the corrected genetic distances