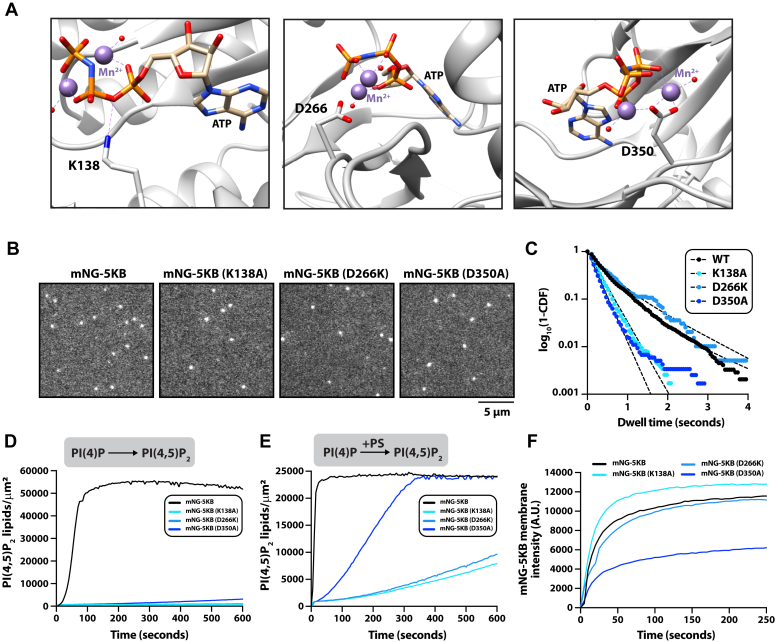
Figure 5.
Deciphering the relationship between PIP5K catalysis and membrane recruitment.A, structure of zPIP5KA (5E3U.pdb) showing the position of residues that are important for catalysis. Residue numbers represent human PIP5KB amino acids that are homologous to the zPIP5KA residues shown in the structure. B, representative TIRF-M images showing the localization of mNG-PIP5KB (WT and mutants). C, single molecule dwell time distributions for mNG-PIP5KB (WT and mutants) measured. See Table 1 for single molecule dwell times (τ1) and statistics. B and C, membrane composition: 98% DOPC, 2% PI(4,5)P2. D and E, lipid kinase activity measured using TIRF-M in the presence of 10 nM PIP5KB (WT and mutants). Production of PI(4,5)P2 was monitored in the presence of 20 nM Cy3-PLCδ. D, membrane composition: 96% DOPC, 4% PI(4)P. E, membrane composition: 78% DOPC, 2% PI(4)P, 20% DOPS. F, bulk membrane recruitment measured by TIRF-M in the presence of 20 nM mNG-PIP5KB (WT and mutants). Membrane composition: 98% DOPC, 2% PI(4,5)P2.