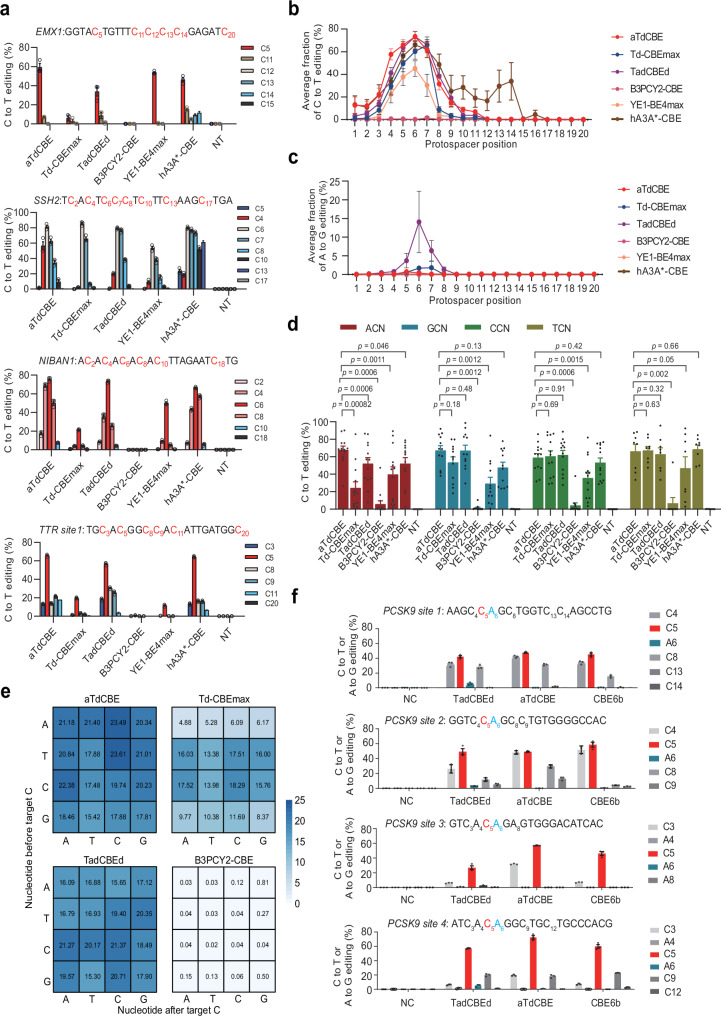
Fig. 3. ATdCBE enables robust genomic base editing in mammalian cells.
a Comparison of C-to-T conversion efficiency for endogenous loci by base editors derived from aTdCBE, Td-CBEmax, TadCBEd, B3PCY2-CBE, hA3A*-CBE and YE1-BE4max in HEK293T cells. b Base editing activity window plots showing mean C-to-T editing at all tested target positions. c Base editing activity window plots showing mean A-to-G editing at all tested target positions. d Comparison of motif preference for 25 endogenous loci by base editors derived from aTdCBE, Td-CBEmax, TadCBEd, B3PCY2-CBE, hA3A*-CBE and YE1-BE4max in HEK293T cells. The editing window of CBEs was primarily concentrated between bases 4–7. Consequently, the analysis focused on preference motifs exhibiting higher editing efficiency within this range. e High-throughput library experiments to evaluate the motif preference of aTdCBE, Td-CBEmax, TadCBEd and B3PCY2-CBE. f Introducing premature termination codons into the PCSK9 coding region using CBEs. The numbers in the grid represent the average editing efficiency of cytosines with corresponding motifs at positions 4–7 for each sgRNA. Data are presented as means ± S.D. Values represent n = 3 independent biological replicates. P-values determined by one-sided Mann-Whitney U-test and adjusted by Benjamini-Hochberg procedure. All of the above base editors contain UGI. NC represents negative control. Source data are provided as a Source Data file.