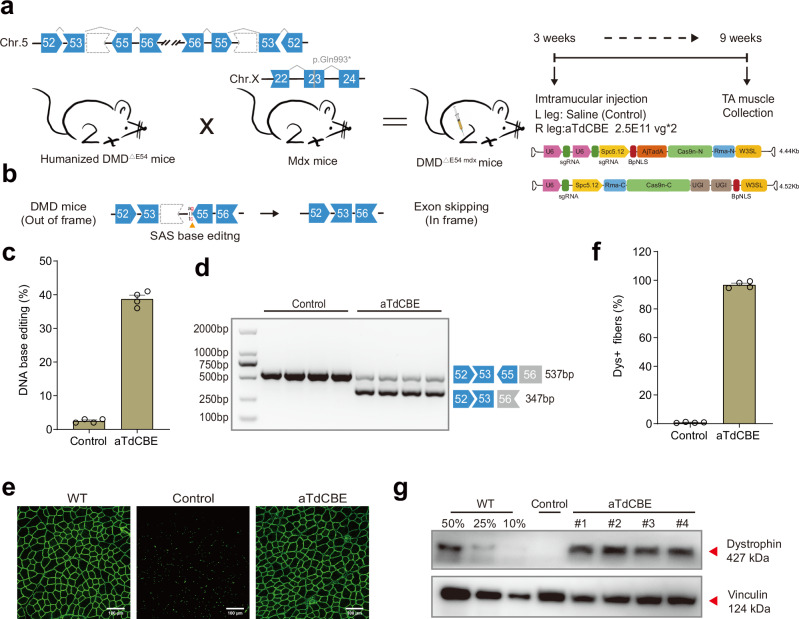
Fig. 4. ATdCBE treatment robustly rescued dystrophin expression in TA six weeks after AAV injection.
a Overview for the in vivo intramuscular (IM) injection of the AAV9-aTdCBE construct into the tibialis anterior (TA) of the right leg of 3-week-old DMDΔ54 mdx mice. The DMDΔE54 mdx mice were derived from the mating of the humanized DMD∆E54 mice with mdx mice. Left leg was injected with saline as a control. Black arrows indicate time points for tissue collection after injection. AjTadA.v2-Y146R, UGIs, Cas9n split by Rma intein and gRNA were packaged into 2 separated AAV particles. A muscle-specific promoter Spc5.12 was used to drive AjTadA-Cas9n-N or Cas9n-C. b Schematic illustrating exon skipping strategies to restore the correct open reading frame (ORF) of the DMD transcript. The shape and color of the boxes representing DMD exons indicate the reading frame. Specifically, the deletion of exon 54 in the DMD gene results in a premature stop codon in exon 55 (depicted in red). The in-frame ORF can be achieved by editing exon 55 splice acceptor sites (SAS) (ag base pair). c The DNA editing efficiency was analyzed after 6-week treatment. d RT-PCR products from muscle of DMDΔE54 mdx mice were analyzed by gel electrophoresis. e Dystrophin (Abcam, ab15277) is shown in green. Scale bar, 100 μm. f Quantification of Dys+ fibers in cross sections of TA muscles. g Western blot analysis of dystrophin (Sigma, D8168) and vinculin (CST, 13901S) expression in TA muscles 6 weeks after injection with AAV-aTdCBE or saline. For comparative analyses, wild-type (WT) mice were derived from crosses between STOCK Tg (DMD) 72Thoen/J mice (#018900) and mdx mice, which carry a c.2977 C > T, p.Gln993* mutation in exon 23 on Chr.X. Dilutions of protein extract from WT mice were used to standardize dystrophin expression (25%, 50% and 75%). Each band (#1-4) represents an individual mouse sample. Vinculin was used as the loading control. Data are represented as mean ± S.E.M (n = 4 independent biological replicates). Each dot represents an individual mouse. Source data are provided as a Source Data file.