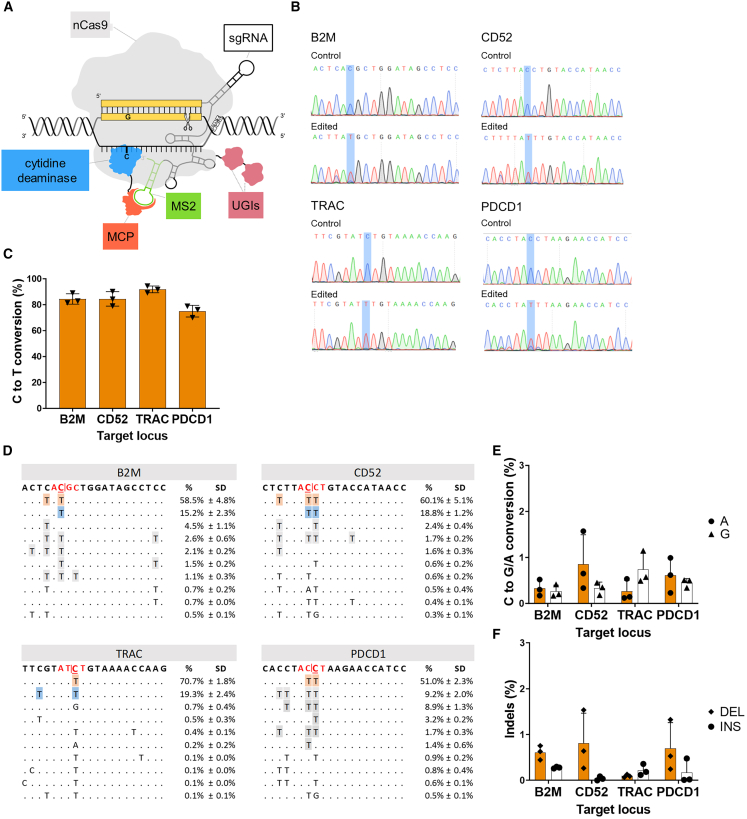
Figure 1.
The Pin-point platform is a highly efficient technology for multiplex editing in T cells
(A) Schematic of the configuration of the Pin-point base editing technology used in this manuscript. An SpCas9 nickase (nCas9-UGI-UGI) binds to the gRNA, the recruiting RNA aptamer (MS2) fused to the gRNA recruits the effector module. The effector module is composed of a cytidine deaminase (rAPOBEC1) fused to the aptamer binding protein (MCP). The recruitment of the deaminase to the target site forms an active complex capable of editing target cytosine residues on the unpaired DNA strand within the CRISPR R-loop. (B) Representative electropherograms from edited and control samples for the four targets following co-delivery of Pin-point mRNAs and four target sgRNAs as analyzed by Sanger sequencing 7 days post electroporation. The target C is highlighted by blue shading. (C) Levels of C to T conversion of the target C at B2M, CD52, TRAC, and PDCD1 loci following co-delivery of Pin-point mRNAs and four target sgRNAs, as analyzed by NGS 7 days post electroporation. Data represented as mean (SEM), n = 3 independent biological T cell donors. (D) Alignment plots showing the top 10 most abundant reads for each of the four targets. The target C is shown in red and underlined. The target splice site is shown in red. (E) Levels of C to G or A conversion of the target C at B2M, CD52, TRAC, and PDCD1 loci following co-delivery of Pin-point mRNAs and four target sgRNAs, as analyzed by NGS. (F) Insertion (INS) and deletion (DEL) frequency at the target C at B2M, CD52, TRAC, and PDCD1 loci following co-delivery of Pin-point mRNAs and four target sgRNAs, as analyzed by NGS. In (E) and (F), data represented as mean (SEM), n = 3 independent biological T cell donors.