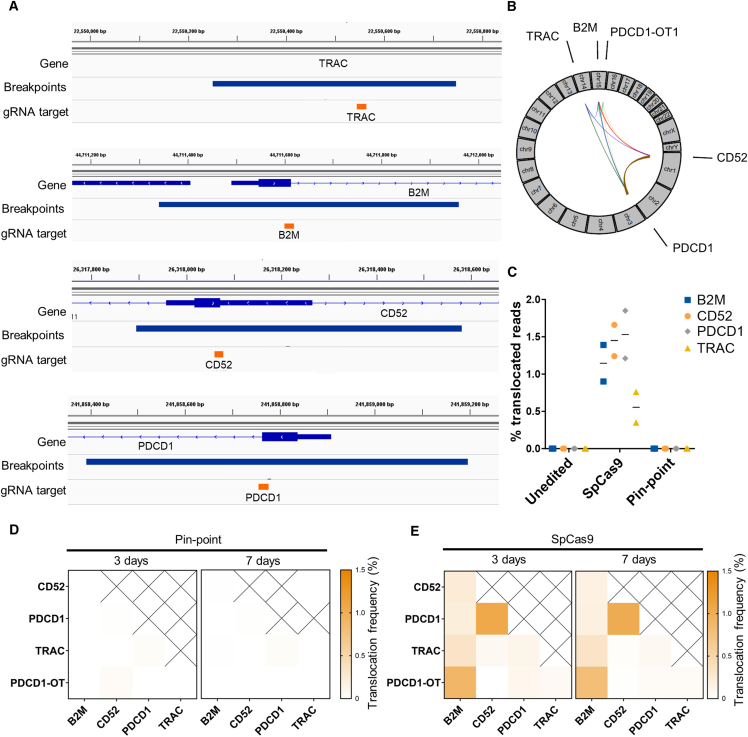
Figure 4.
Assessment of translocations following multi-gene editing
(A) gRNA binding sites (orange) and genomic regions spanning the locations of SpCas9-induced translocation breakpoints identified by the DRAGEN Structural Variant (SV) Caller (blue bars) at each target gene locus. Genomic regions include confidence intervals around each breakpoint. (B) Circos plot of interchromosomal translocations involving gRNA target sites identified by MANTA in the SpCas9 multi-edited samples. Thickness of lines joining genomic loci is proportional to the translocation frequency. (C) Percentage of Capture-seq reads marked as translocations mapping to each sgRNA target site. (D and E) Average frequencies of the two outcomes of each predicted reciprocal translocation quantified by ddPCR 3 or 7 days post editing with either (D) the Pin-point platform or (E) SpCas9. For (B)–(E) mRNAs encoding either the Pin-point platform or SpCas9 were delivered with four targeting sgRNAs. Control is mock electroporated T cells without RNA. Samples were analyzed 3 days post electroporation unless specified otherwise. n = 2 independent T cell donors.