Figure 5.
Effect of the Pin-point platform on RNA editing and transcription
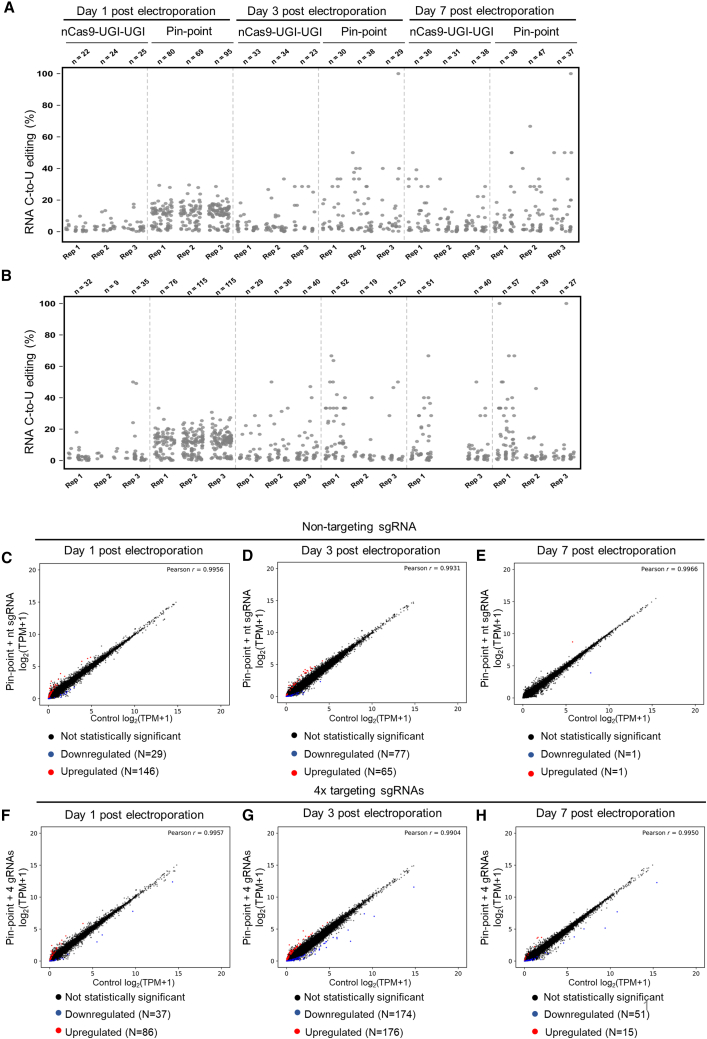
(A and B) RNA C to U editing assessed by transcriptome sequencing in primary human T cells that were electroporated with Pin-point (nCas9-UGI-UGI and rAPOBEC1-MCP) or nCas9-UGI-UGI only mRNAs and the four targeting sgRNAs against B2M, CD52, TRAC, and PDCD1 genes (A) or a scrambled non-targeting sgRNA (B). Each dot represents one editing event. The total number of editing events is indicated above each replicate per condition. (C–H) Scatterplots of gene expression levels (log2 transformed TPM +1, TPM with a pseudocount of one added before log transformation) in primary human T cells electroporated with Pin-point mRNAs and either a scramble non-targeting sgRNA (C, D, and E) or the four targeting sgRNAs (TRAC, B2M, CD52, PDCD1) (F, G, and H) compared with control cells that received the pulse electroporation only (x axis). DESeq2 analysis was performed on total mRNA collected at days 1 (C and F), 3 (D and G), and 7 (E and H) post electroporation and was used to identify upregulated and downregulated genes. Upregulated or downregulated genes (p < 0.05) with absolute log2 fold change ≥1.5 in gene expression (represented as log2 transformed TPM +1) marked red and blue, respectively. r indicates the Pearson correlation coefficient, calculated for log-transformed values on all genes.