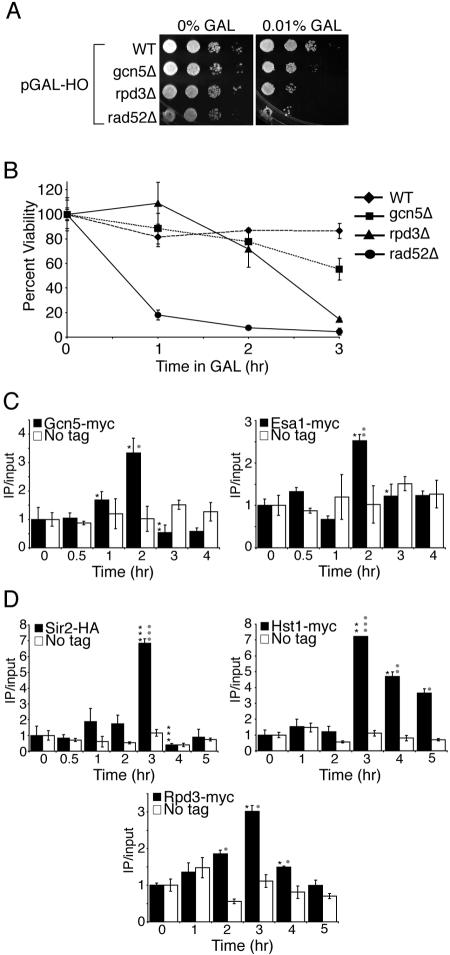
FIG. 3.
Gcn5, Esa1, Sir2, Hst1, and Rpd3 may act to modify histones during homologous recombination. A. Sensitivity to prolonged HO induction. Serial dilution analysis of strains BAT021 (wild type [WT]), BAT019 (gcn5Δ), BAT020 (rpd3Δ), and BAT022 (rad52Δ), containing plasmid pGAL-HO-URA3 onto plates with and without galactose (GAL). B. Sensitivity to transient induction of HO endonuclease. Viability analysis of the strains used in panel A following transient exposure to HO endonuclease performed as described in the legend to Fig. 1B. C. Gcn5 and Esa1 are recruited to the HO lesion. ChIP analysis of strains Z1466 (Gcn5-MYC), Z1346 (Esa1-Myc), and Z1256 (no tag) containing a pGAL-HO-URA3 plasmid where galactose was added after time zero and glucose was added after 2 h to induce and repress the HO endonuclease. Real-time PCR analysis and quantification was performed as described in the legend to Fig. 2 using primers 0.6 kb to the right of the HO site and an SMC2 internal primer pair for normalization. D. Sir2, Hst1, and Rpd3 are recruited to the HO lesion. ChIP analysis of strains BAT035 (Sir2-HA) and BAT009 (No tag), Z1346 (Hst1-MYC) and Z1256 (No tag), and Z1347 (Rpd3-MYC) and Z1256 (No tag) containing a pGAL-HO-URA3 plasmid as described for panel C.