Figure 1.
Pseudobulk and single-cell-level differentially expressed gene analyses suggested escape from XCI across immune cells
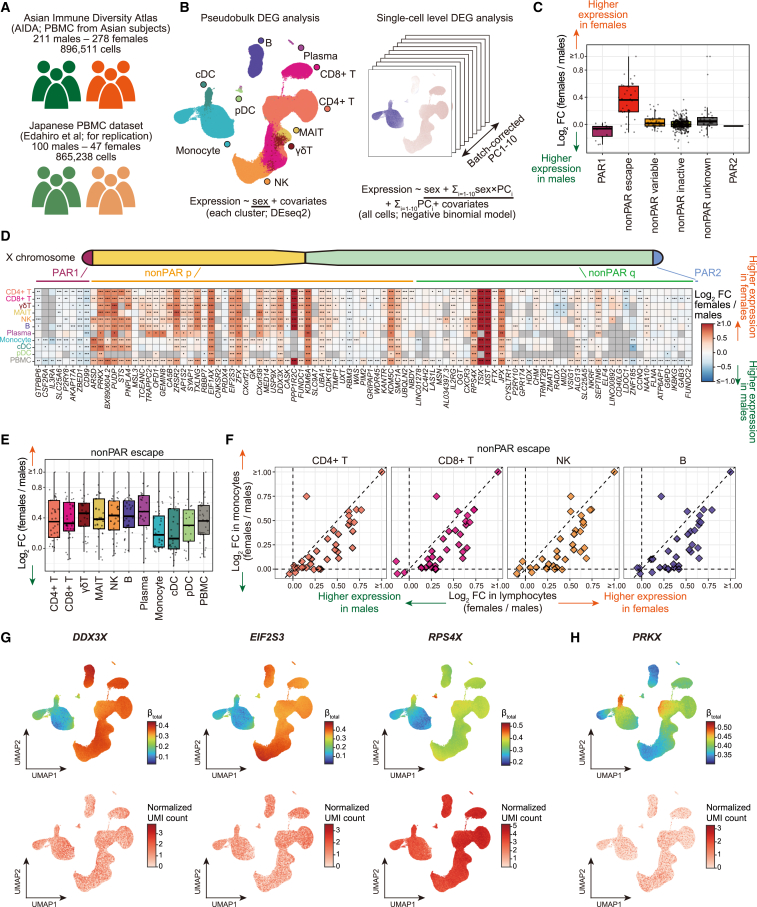
(A) The scRNA-seq datasets used in this study.
(B) The DEG analysis methods used in this study (STAR Methods, Data S1).
(C) A boxplot represents log2 fold changes in the gene expression between sexes. Genes are grouped according to the XCI status annotated in the previous study.2
(D) A heatmap represents differential gene expression between sexes. The colors of the tiles represent log2 fold changes in the gene expression between sexes. Only genes that satisfied Bonferroni-corrected significance thresholds at least in one cell type are shown. ∗p < 0.05. ∗∗Per-cell-type false discovery ratio (FDR) <0.05. ∗∗∗Bonferroni-corrected p < 0.05.
(E) A boxplot represents log2 fold changes of the escapee gene expression between sexes across cell types.
(F) Scatterplots represent pairwise comparisons of the log2 fold changes of the escapee gene expression between sexes. The y axes represent the log2 fold changes in monocytes and the x axes represent the log2 fold changes in lymphocytes. The dashed lines represent x = 0, x = y, and y = 0.
(G and H) UMAPs represent the per-cell effect sizes of the sex in the single-cell-level DEG analysis calculated as a sum of the effect sizes of sex and sex × batch-corrected PCs (STAR Methods, top) and gene expression (bottom). Genes that show a stronger degree of escape in lymphocytes than monocytes (G) and other patterns of heterogeneity of effect sizes (H) are indicated. The p values for the interaction between sex and batch-corrected PCs were <1 × 10−200 (G) and 1.5 × 10−12 (H). DEG, differentially expressed genes; PC, principal component; PAR, pseudoautosomal region; PBMC, peripheral blood mononuclear cells; scRNA-seq, single-cell RNA-seq; UMAP, uniform manifold approximation and projection; XCI, X chromosome inactivation.