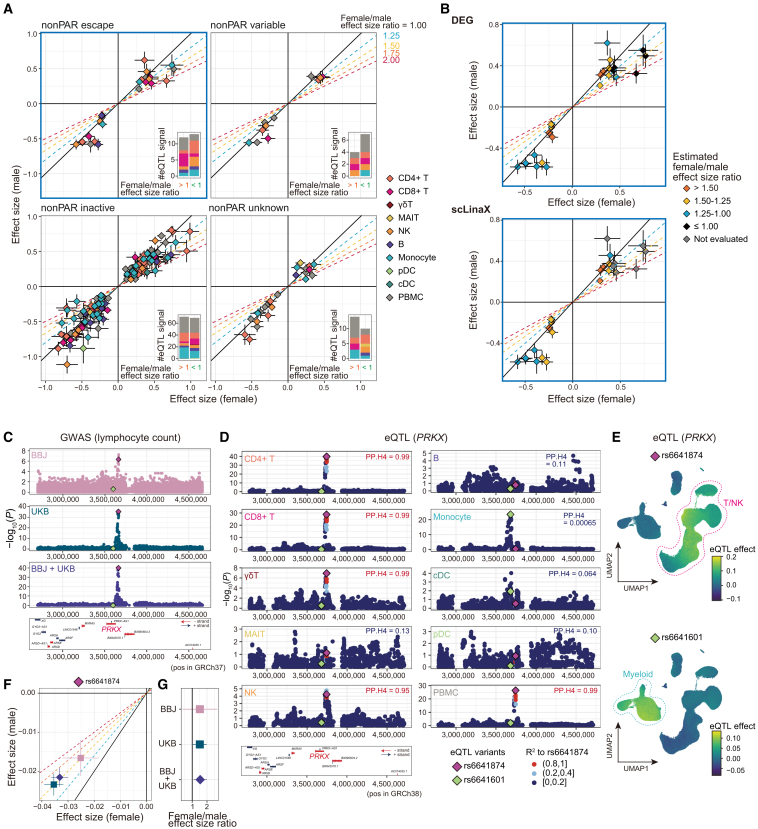
Figure 6.
Detection of differential effect sizes between sexes in the genotype-phenotype association analysis
(A) The effect sizes of the significant eQTL signals (p < 5 × 10−8) in the female-only (x axis) and male-only (y axis) analyses, separately for each XCI status. The error bars indicate standard errors. The color of the plots indicates the cell type in which the eQTL signals are identified. The oblique lines correspond to the female/male effect size ratios described in the plots. The bar plots in the lower right of each plot indicate the number of eQTL signals that have larger effect sizes in females (left) and males (right).
(B) Scatterplots for escapee genes (A, upper left) are colored according to the estimated female/male effect size ratio based on the DEG analysis (top) and scLinaX analysis (bottom). Genes that were not evaluated in the scLinaX analyses are colored gray.
(C) The association between PRKX gene loci and lymphocyte counts in the BBJ analysis, the UKB analysis, and the BBJ + UKB meta-analysis. The rs6641874 (top variant in the BBJ + UKB meta-analysis and T cells eQTL analysis) and rs6641601 (top variant in the monocytes eQTL analysis) are colored purple and green, respectively. Genes located around the PRKX gene region are indicated at the bottom of the plots.
(D) Locus plots for the eQTL analysis of the PRKX gene across cell types. R2, a measure of linkage disequilibrium (LD) to the rs6641874, is indicated by the color of the dots. Results of the colocalization analyses (PP.H4) with lymphocyte count GWASs in BBJ are indicated in the upper right of the plots.
(E) UMAPs represent the per-cell eQTL effect sizes of the variants in the single-cell-level eQTL analysis calculated (STAR Methods). Associations for PRKX genes rs6641874 (top) and rs6641601 (bottom) are indicated. The p values for the interaction between genotypes and batch-corrected PCs were 2.7 × 10−91 (top) and 2.4 × 10−51 (bottom).
(F) The effect sizes of the rs6641874 in the female-only (x axis) and male-only (y axis) lymphocyte count GWAS analyses in each cohort. The error bars indicate standard errors.
(G) The female/male effect size ratios of the rs6641874 in the lymphocyte count GWAS analyses in each cohort. The error bars indicate 95% CI.