Figure 3.
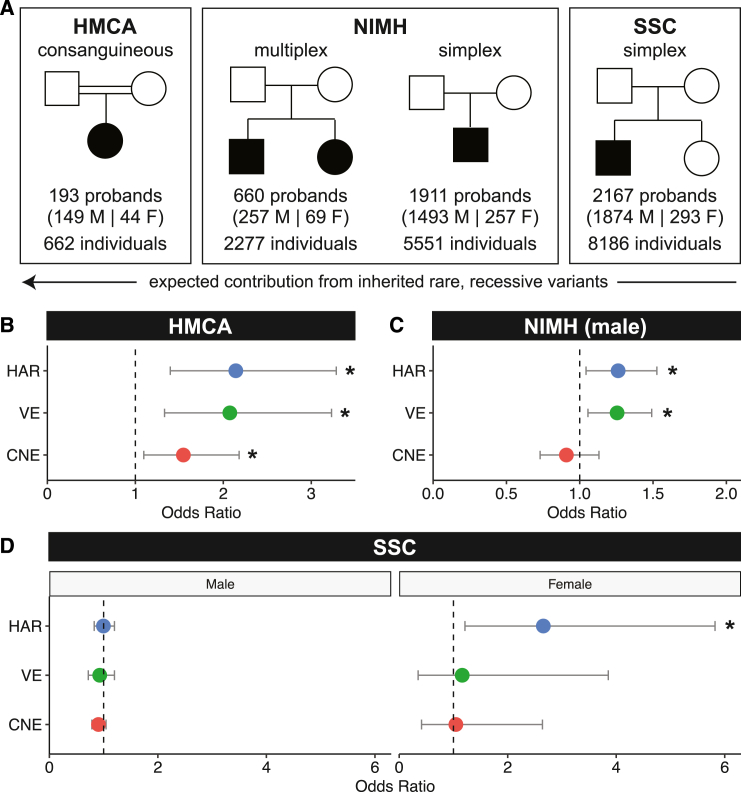
Contribution of rare, recessive variants in HARs, VEs, and CNEs to ASD varies across cohorts based on family structure
(A) ASD cohorts.
(B) In the HMCA cohort, cases are enriched for rare, recessive variants in HARs (adjusted p = 0.0014), VEs (adjusted p = 0.0038), and CNEs (adjusted p = 0.0412) at allele frequency (AF) < 0.005.
(C) In the NIMH cohort, male cases are enriched for rare, recessive variants in HARs (adjusted p = 0.0495) and VEs (adjusted p = 0.0297) at AF < 0.001.
(D) In the SSC cohort, female cases are enriched for rare, recessive variants in HARs (adjusted p = 0.0438) at AF < 0.005.
All analyses were done on conserved bases. Odds ratios and 95% confidence intervals were calculated as previously described,49 and p values comparing odds ratios were calculated using z values assuming deviation from a normal distribution. Full details of statistical analyses are in STAR Methods.