Fig. 1.
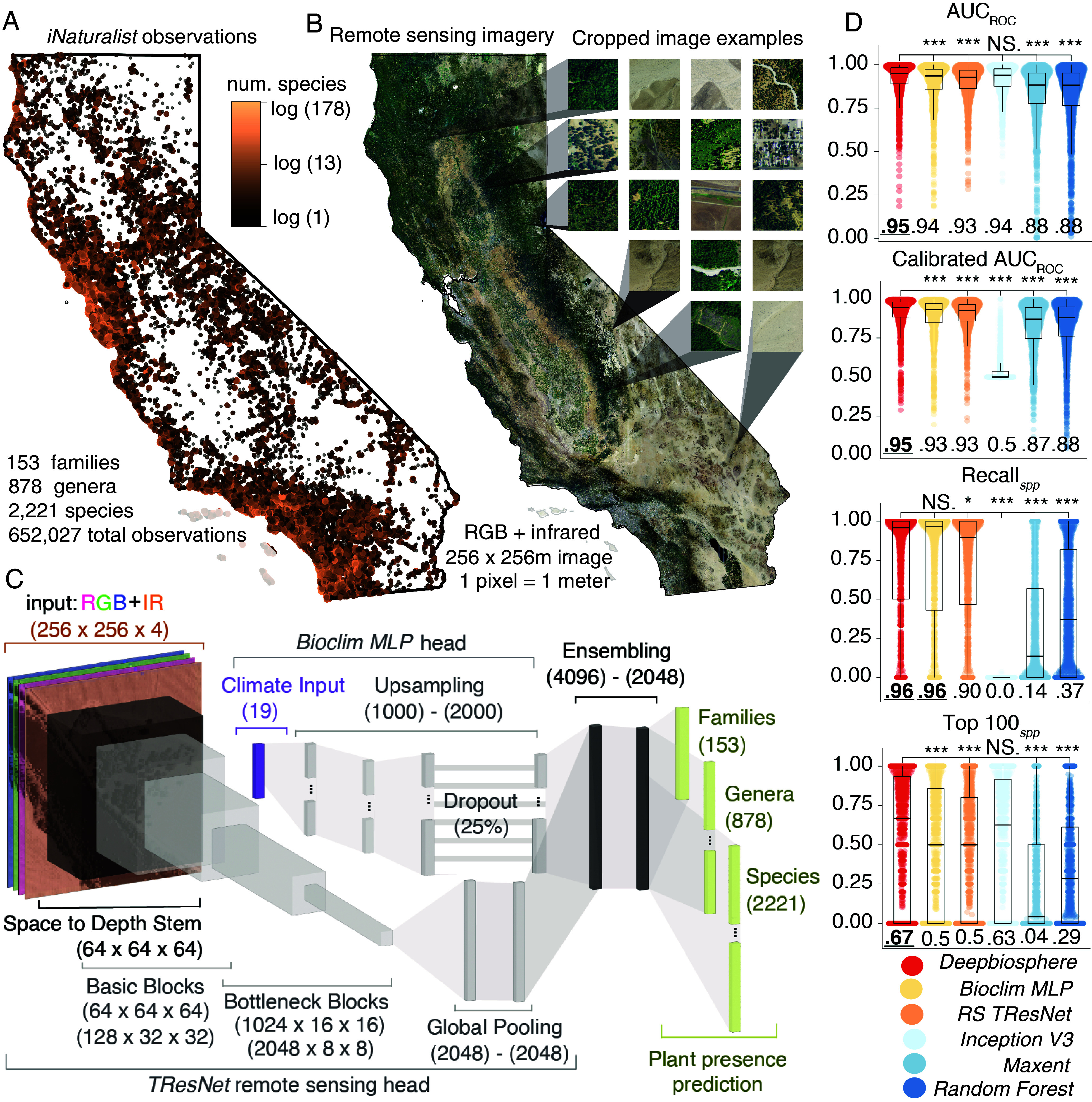
Training a deep neural network to predict the presence of thousands of plant species. (A) Map of over 650,000 iNaturalist observations for 2,221 plant species we curated to train plant species distribution models (SDMs) (24). (B) To train the CNNs, species observations were linked to 256 × 256 m remote sensing images cropped from 2012 NAIP data (16), along with climate variables (31) (SI Appendix, Fig. S2). (C) Deepbiosphere architecture, which combines a residual CNN (TResNet) (27) trained using remote sensing imagery with a multilayered perceptron (MLP) network (32) trained using climate variables to predict plant families, genera, and species (SI Appendix, Table S3). Layer types and dimensions are annotated for each section of the neural network. (D) Comparison of Deepbiosphere’s performance to common climate-based SDMs including Maxent, Random Forest (33), as well as the Bioclim MLP head trained with just climate, the remote sensing TResNet head trained with just NAIP data, and an Inception V3 model from previous work (34). Metrics are reported per-species for the 1,541 species shared between the uniform split of the training and testing set, with the median score annotated on each boxplot. spp = per-species, AUC = area under the curve; ROC = receiver operating characteristic curve; RS = remote sensing. Stars indicate results from unpaired student’s t test, with *** indicating a P-value < 10−3, * indicating a P-value < 10−1, and NS. indicating a nonsignificant P-value of > 0.1.
