Abstract
Background
Bacillus cereus is a Gram-positive, spore-forming bacterium that produces a spectrum of effectors integral to bacterial niche adaptation and the development of various infections. Among those is EsxA, whose secretion depends on the EssC component of the type VII secretion system (T7SS). EsxA’s roles within the bacterial cell are poorly understood, although postulations indicate that it may be involved in sporulation. However, the T7SS repertoire in B. cereus has not been reported, and its functions are unestablished.
Methods
We used the type strain, B. cereus ATCC14579, to generate ΔessC mutant through homologous recombination using the homing endonuclease I-SceI mediated markerless gene replacement. Comparatively, we analyzed the culture supernatant of type strain and the ΔessC mutant through Liquid chromatography-tandem mass spectrometry (LC-MS/MS). We further generated T7SSb-specific gene mutations to explore the housekeeping roles of the T7SSb-dependent effectors. The sporulation process of B. cereus ATCC14579 and its mutants was observed microscopically through the classic Schaeffer-Fulton staining method. The spore viability of each strain in this study was established by enumerating the colony-forming units on LB agar.
Results
Through LC-MS/MS, we identified a pair of nearly identical (94%) effector proteins named EsxA belonging to the sagEsxA-like subfamily of the WXG100 protein superfamily in the culture supernatant of the wild type and none in the ΔessC mutant. Homology analysis of the T7SSb gene cluster among B. cereus strains revealed diversity from the 3’ end of essC, encoding additional substrates. Deletions in esxA1 and esxA2 neither altered cellular morphology nor growth rate, but the ΔesxA1ΔesxA2 deletion resulted in significantly fewer viable spores and an overall slower sporulation process. Within 24 h culture, more than 80% of wild-type cells formed endospores compared to less than 5% in the ΔesxA1ΔesxA2 mutant. The maximum spore ratios for the wild type and ΔesxA1ΔesxA2 were 0.96 and 0.72, respectively. Altogether, these results indicated that EsxA1 and EsxA2 work cooperatively and are required for sporulation in B. cereus ATCC14567.
Conclusion
B. cereus ATCC14579 possesses two nearly identical T7SSb-dependent effectors belonging to the sagEsxA-like proteins. Simultaneous deletion of genes encoding these effectors significantly delayed and reduced sporulation, a novel finding for EsxA.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03492-1.
Keywords: Bacillus cereus, Type VIIb secretion system, EsxA, Sporulation
Introduction
Bacillus cereus, a Gram-positive, spore-forming bacterium belonging to the Firmicutes phylum, has been primarily associated with food poisoning. But recently, some strains have been implicated in severe infection, including bacteremia [1, 2], while others have evolved to produce lethal anthrax-like disease [3]. B. cereus has been considered a neglected pathogen, but studies are now unraveling the evolution of its virulence mechanisms. The ability to express a variety of virulence factors, including pore-forming toxins, hemolysins, enterotoxins, proteases, and phospholipases, is central to its ability to cause infections in humans and animals as well as colonizing the environment [4].
Among the Firmicutes bacterial mechanisms that facilitate their virulence is the secretion system for the passage of toxigenic materials. Like other Gram-positive bacteria, B. cereus is expected to have a type VII secretion system (T7SS) possessing a specialized transmembrane nanomachinery, EssC, belonging to the FtsK/SpoIIIE ATPase protein family. The T7SS was first discovered in Mycobacterium tuberculosis [5], and recent studies on Gram-positive bacteria have shown limited homology to the mycobacterial T7SS components, which led to the sub-classification of T7SS into T7SSa (Actinobacteria) and T7SSb (Firmicutes) [6, 7]. Despite a limited structural similarity, T7SS share highly similar substrates, including the early secretory antigenic targets (ESAT) A (EsxA) and B (EsxB), containing a conserved Trp–Xaa–Gly (WXG) central sequence motif. EsxA and EsxB are small acidic proteins of the WXG100 superfamily, which have shown to be strong cytotoxic T-cell antigens [8, 9] and possess pore-forming activity [10]. EsxA alone or in combination with EsxB causes dose-dependent pore-formation in host cell membranes, which could be crucial for the translocation of other bacterial virulence factors into the host [10, 11]. Recently, studies in Staphylococcus aureus have shown that secretion of these effector proteins requires a functional EssC [7, 12]. Studies have also established that EsxB alone binds via its C-terminal secretion signal sequence to an empty pocket on the EssC C-terminal, accompanied by an increase in ATPase activity. Surprisingly, substrate binding does not activate EssC allosterically but by stimulating its multimerization, forming a hexameric pore necessary for the secretion of T7SS-dependent effectors [13]. These findings suggest that WXG100 proteins are virulence factors outside the cell and may have other housekeeping roles since they are also found among non-pathogenic organisms [14, 15].
Further, T7SS genes encoding additional WXG100-like effectors, EsxC and EsxD, have been identified in S. aureus, and the polymorphic LXG toxin among Firmicutes and their functions have been characterized (Fig. 1) [8, 16, 17]. The TIGR04197 proteins have recently been remotely linked to the T7SS based on phylogenetic profiling as a putative T7SS effector. In addition, a site-specific mutation in the TIGR04197 encoding gene abrogated LXG effector export, suggesting its significance in LXG effector secretion by the T7SSb apparatus in Streptococcus intermedius [18]. Despite many studies on T7SS in various species of medical importance, the repertoire of the secretion system in B. cereus has not been reported and is always inferred from other Firmicutes members. Among the closely related Bacillus species, the T7SS has been established in B. subtilis (Fig. 1) [19, 20] and B. anthracis [21].
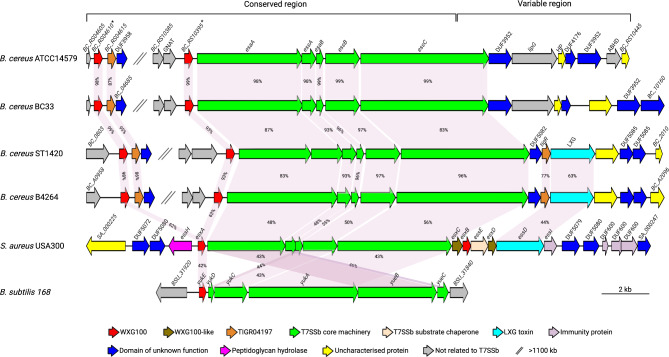
Fig. 1.
Schematic representation of the T7SSb gene cluster in B. cereus ATCC14579 (GenBank: NC_004722.1). The putative WXG100 protein-encoding genes, BC_RS04610 and BC_RS10395, are indicated with asterisks. Homology analysis of the corresponding T7SSb gene clusters in different B. cereus strains S. aureus USA300 and B. subtilis 168 are also shown. The same color indicates related genes, and percentage identities for each gene, with reference to B. cereus ATCC14579, are shown in pink-shaded areas. Genes at the 5’ end of the essC are highly conserved, while variation is observed in the 3’ end
Like EssC, SpoIIIE belongs to the FtsK/SpoIIIE ATPase protein family, but it is indispensable for DNA translocation from the mother cell to the apically located forespore during sporulation [22]. B. cereus possesses both EssC and SpoIIIE for the T7SSb and spore formation, respectively. SpoIIIE acts as a checkpoint that prevents septal membrane fusion until the completion of chromosome translocation via the aqueous DNA-conducting hexameric pole. Both proteins are multidomain with high homology in the substrate-binding C-terminal motor domain, and recruitment of multiple factors precedes multimerization [13, 23, 24]. However, it is unknown whether an EssC substrate can bind to another FtsK/SpoIIIE protein, such as SpoIIIE, and affect its function(s).
Therefore, this study first sought to identify the T7SSb-dependent effectors and establish the T7SSb gene locus in B. cereus. Relying on data from the well-studied T7SSb in S. aureus USA300, we generated a B. cereus ATCC14579 ΔessC mutant to identify T7SSb-dependent effectors. Here, we performed a comprehensive proteomic analysis of the culture supernatant for B. cereus ATCC14579 and its ΔessC mutant and identified two nearly identical EsxA proteins, EsxA1 and EsxA2. Secondly, we explored the influence of the identified effectors in the phenotypic characteristics of B. cereus ATCC14579 after generating single and double mutations for the T7SSb effectors genes. We established that B. cereus ATCC14579 ΔesxA1ΔesxA2 displayed a significantly decreased sporulation phenotype compared to the wild type.
Materials and methods
Bacterial strains and growth conditions
For routine culture, B. cereus strains were grown in Brain Heart Infusion (BHI) medium (Difco, USA) at 37 °C (shaking at 155 rpm for liquid cultures) and Escherichia coli strains in Luria-Bertani (LB) medium (Difco, USA). The bacterial strains and plasmids used in this study are shown in Table 1. Cultures for protein mass spectrometry were done in minimal medium (M9), 2% glucose, 50 mM Na2HPO4, 25 mM KH2PO4, 10 mM NaCl, 20 mM NH4Cl, 1 mM MgSO4·7H2O, 0.1 mM CaCl2, 10 mM glutamic acid, 1.4 mM threonine, 0.6 mM leucine, 2.5 mM valine, 1.4 mM histidine and 0.3 mM methionine at 37 °C. When needed, antibiotics were added to LB at final concentrations of 250 µg/ml (B. cereus) or 100 µg/ml (E. coli) spectinomycin, 60 units/ml polymyxin, 50 µg/ml kanamycin, and 100 µg/ml ampicillin. For sporulation assays, we used 0.8% Nutrient Broth (Difco, USA), 1.2% (w/v) MgSO4·7H2O, 10% (w/v) KCl, 1 mM Ca(NO3)2, 0.01 mM MnCl2·6H2O, and 1 µM FeSO4·7H2O (pH = 7.6) [25] under aerobic conditions at 30 °C with shaking at 200 rpm.
Table 1.
Strains and plasmids used in this study
| Strain | Genotype or Description | Source |
|---|---|---|
| B. cereus | ||
| ATCC14579 | Wild type | [28] |
| ΔessC (essC gene deleted mutant) | This study | |
| ΔesxA1 (esxA1 gene deleted mutant) | This study | |
| ΔesxA2 (esxA2 gene deleted mutant) | This study | |
| ΔtigR (tigR gene deleted mutant) | This study | |
| ΔesxA1ΔesxA2 (esxA1 and esxA2 genes deleted mutant) | This study | |
| ΔesxA1ΔesxA2 + pGFP78::esxA1 (ΔesxA1ΔesxA2 mutant complimented with its native esxA1 gene) | This study | |
| ΔesxA1ΔesxA2 + pGFP78::esxA2 (ΔesxA1ΔesxA2 mutant complimented with its native esxA2 gene) | This study | |
| ΔesxA1ΔesxA2 + pGFP78 (ΔesxA1ΔesxA2 mutant complimented with pGFP78 plasmid) | This study | |
| Wild type + pGFP78 (Wild type complimented with pGFP78 plasmid) | This study | |
| E. coli | ||
| SM10 | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Km) | [29] |
| S17-1 | thi, pro, hsdR, RP4-2 Tc::Mu Km::Tn7 (Tp Sm) | [29] |
| DH10β (C3019H) | Δ(ara-leu) 7697 araD139 fhuA ΔlacX74 galK16 galE15 e14- Φ80dlacZΔM15 recA1 relA1 endA1 nupG rpsL (StrR) rph spoT1 Δ(mrr-hsdRMS-mcrBC) | New England BioLabs |
| Plasmids | ||
| pRP1028 | Bacillus and E. coli shuttle vector with turbo-rfp gene and an I-SceI recognition site; SpeR | [26] |
| pRP1099 | Vector with turbo-AmCyan and I-SceI genes; KanR | [26] |
| pGFP78 | Shuttle vector for Bacillus and E. coli containing gfp gene and F78 promoter; AmpR, TetR | [27] |
| pYF07 | pRP1028 with upstream and downstream regions of essC | This study |
| pHKK100 | pRP1028 with upstream and downstream regions of esxA1 | This study |
| pHKK102 | pRP1028 with upstream and downstream regions of esxA2 | This study |
| pHKK104 | pRP1028 with upstream and downstream regions of tigR | This study |
| pHKK114 | pGFP78 Δgfp with esxA1 | This study |
| pHKK115 | pGFP78 Δgfp with esxA2 | This study |
SpeR, KanR, TetR, and AmpR stand for spectinomycin, kanamycin, tetracycline, and ampicillin resistance, respectively
Construction of plasmids and gene knockout procedure
To construct the essC-knockout mutant strain, the upstream (US) and downstream (DS) homologous arms of B. cereus ATCC14579 were inserted into plasmid pRP1028 between the BanII-SacI and BsaI sites to construct the pYF07. The US and DS fragments were generated via PCR using the primers indicated in Table 2. PCR and sanger sequencing further verified the resulting plasmids using appropriate primers. The same procedure was used to construct pHKK100, pHKK102, and pHKK104 plasmids. The allelic exchange system used in this study was developed based on the homing endonuclease I-SceI mediated markerless gene replacement method established in B. anthracis [26] with minor variations. We used the biparental mating method, eliminating the use of the helper strain (E. coli SS1827). We used E. coli SM10 and S17-1, which have tra genes encoding factors that can mobilize oriT-containing plasmids and integrate into chromosomes, thus making biparental mating possible. E. coli SM10 was used for pRP1028 derivatives conjugative transfer, and E. coli S17-1 was used to transfer pRP1099. Confirmation of gene deletion was done using PCR and sanger sequencing. This allelic exchange procedure is illustrated in Figure S1. The same method was used to create ΔesxA1, ΔesxA2, and ΔtigR mutants of B. cereus ATCC14579. To construct the double mutant, we used B. cereus ΔesxA1 and E. coli SM10 harboring pHKK102 and followed the same allelic exchange procedure from step 4 (Fig. S1).
Table 2.
Primers used in this study
| Primer name | Sequence (5’–3’)a | Application |
|---|---|---|
| essC_US_F | cttgagctcctagcggccgcaagaaatgagaaattaagactacttcgg | For construction of ΔessC mutant |
| essC_US_R | ttaatttcagaattccatccgtgttcactgctc | |
| essC_DS_F | acacggatggaattctgaaattaaagaagaaagctaaggtg | |
| essC_DS_R | gtacaggtctccggccgctctgcgttaaatttatcctgtaatgtc | |
| esxA1_US_F | tcctagcgatgtgtgactatattggctgtttttatccaga | For construction of ΔesxA1 mutant |
| esxA1_US_R | gtatgacaattttttcatttcccctttaatacataatgagcaaaatataaattgg | |
| esxA1_DS_F | gggaaatgaaaaaattgtcatactctatataacaagcatttcaaatgagc | |
| esxA_1_DS_R | tctccggccttaacttttgatttacttgattgatttgtttttcaatatcttgact | |
| esxA2_US_F | gcttgagcggatggatattaataggggaaaatacagactatttttct | For construction of ΔesxA2 mutant |
| esxA2_US_R | ctttttatctgtctattaaatcccccttaaaattaaaaaatagtaaaattaaccaataatca | |
| esxA2_DS_F | ggggatttaatagacagataaaaagaaatatataatttagccatgcaacc | |
| esxA2_DS_R | cgctaggattctgcctcagctttcaaatcct | |
| tigR_US_F | agcttgagcgttagtttggctgtagtagatattatcgttgcgaat | For construction of ΔtigR mutant |
| tigR_US_R | ggttgtcacacactcaaatcctttctctaattttcagaacatg | |
| tigR_DS_F | atttgagtgtgtgacaacctacgtaaaggtaaaatagtaagagga | |
| tigR_DS_R | gccgctaggactaatatttcttgttcaattacatctgtggaagc | |
| Dx_esxA1_F | ccaatttatattttgctcattatgtattaaagggg | PCR and sanger sequencing confirmation of esxA1 deletion |
| Dx_esxA1_R | ctttgaaattgtcccatcactcaaatcctttctc | |
| Dx_tigR_F | ggctcatttgattgcatgcccaac | PCR and sanger sequencing confirmation of tigR deletion |
| Dx_tigR_R | cctgaatgtgaatcgcggattgatttcggtc | |
| Dx_esxA2_F | ggtttgcctttattttatagtagatttatagagtgtgc | PCR and sanger sequencing confirmation of esxA2 deletion |
| Dx_esxA2_R | cgactgcgagtctatctaatgaaagacttcgtatc | |
| esxA1_F_C | gctctagacaatttatattttgctcattatgtattaaagggg | For construction of esxA1 complimentary strain |
| esxA1_R_C | cccaagcttgctcatttgaaatgcttgttatatagagtatgac | |
| esxA2_F_C | gctctagaggtttgcctttattttatagtagatttatagagtgtgc | For construction of esxA2 complimentary strain |
| esxA2_R_C | cccaagcttcgactgcgagtctatctaatgaaagacttcg |
a Restriction sites are italicised
Construction of complimentary strains
To complement the B. cereus ΔesxA1ΔesxA2 mutation, a GFP plasmid, pGFP78, an Escherichia coli–Bacillus subtilis shuttle vector carrying a constitutive 78 promoter-controlled GFP gene was used. The GFP open reading frame was removed from this vector by digestion with XbaI and HindIII [27]. B. cereus ATCC14579 genome was used to amplify esxA1 and esxA2 using specific primers in Table 2, and each was ligated to the XbaI and HindIII sites of the linearized plasmid to generate each respective complementation plasmid. The orientation and sequence of PCR fragments were confirmed by sanger sequencing. The recombinant plasmids were each electroporated into B. cereus ΔesxA1ΔesxA2 producing B. cereus ΔesxA1ΔesxA2 + pGFP78::esxA1 and B. cereus ΔesxA1ΔesxA2 + pGFP78::esxA2 mutants. Successful transformants were selected on LB agar plates supplemented with 10 µg/ml tetracycline and further confirmation by sanger sequencing. In addition, we complemented B. cereus ATCC14579 and the ΔesxA1ΔesxA2 mutant with pGFP78 for growth in tetracycline. All strains constructed in this study are listed in Table 1.
Liquid chromatography-tandem mass spectrometry
B. cereus ATCC14579 and its ΔessC mutant were each grown in 2.5 ml LB at 37 °C for 18 h, and then cells were pelleted and washed three times in M9. Resuspended cells were incubated in M9 with start OD600 = 0.05 at 37 °C until mid-log phase (∼5 h). The cells were pelleted and filtered the culture supernatant using a 0.45 μm membrane filter (Millipore, Ireland). The supernatant was purified using methanol-based C8 reverse-phase liquid chromatography (Strata™ Phenomenex, USA). The purified protein mixture then underwent alkylation, tryptic digestion, and further peptide purification using Pierce™ C18 spin columns (ThermoFisher Scientific, USA) following the manufacturer’s protocol. The peptide mixtures were analyzed using the HPLC liquid phase system EASY nLC1000 (ThermoFisher Scientific, USA). They were loaded onto a C18 column (3 μm particle diameter, 0.075 mm x 120 mm, Nikkyo Technos, Japan) at a 300 nl/min flow rate controlled by intelliflow technology over 81 min. The HPLC was coupled online via a nanoelectrospray ion source to a hybrid ion trap quadrupole-Orbitrap (LTQ–OrbitrapXL; ThermoFisher Scientific) tandem mass spectrometer (LC-MS/MS) [30]. The LC − MS/MS.RAW files were searched against B. cereus ATCC 14,579 protein FASTA file database and annotated the peptides using the Sequest HT algorithm in Proteome Discoverer 1.4.1.14 (DBVersion:79) (Thermo Fisher Scientific, Waltham, MA). The reference strain B. cereus BC33 (GenBank: CP072774.1) was also used as a database to validate the peptide annotations from the B. cereus ATCC14579 database. Search parameters were set to two missed trypsin cleavage sites, and a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.06 Da were utilized. The search included carbamidomethylation of cysteine as a static modification and amine carbamylation and methionine oxidation as dynamic modifications, with a maximum of four modifications per peptide. The false discovery rate for peptide-spectrum matches (PSMs) was set at 1% maximum using a target-decoy PSM validator [31, 32].
B. cereus T7SSb-dependent effectors and gene cluster
Sequences of annotated proteins in the mass spectrometry data were obtained from the GenBank files and analyzed for motifs found among known T7SS substrates using the GenomeNet platform [33] to validate their identity. Further bioinformatic searches were conducted using web resources NCBI BLASTp [34] (available at https://blast.ncbi.nlm.nih.gov/Blast.cgi) to obtain the best hits among Firmicutes for CLUSTAL O alignment. Furthermore, genes encoding the candidate T7SSb effectors were obtained for homology and neighborhood analysis against the well-studied and established T7SSb in S. aureus USA300 (GenBank: CP092052.1) and B. subtilis 168 (GenBank: AL009126.3). This analysis was used to identify the T7SSb core machinery encoding genes, and the output data was imported into genoPlotR [35] to construct the gene cluster. Additionally, a comparative analysis of genome sequences was performed to detect T7SSb in the reference strain, B. cereus BC33 (GenBank: CP072774.1), and the more virulent strains, B. cereus ST1420 ( GenBank: AP022975.1) and B. cereus B4264 (GenBank: CP001176.1). CLUSTAL O version 1.2.4 multiple sequence alignment was used to calculate the T7SSb nucleotide sequence similarity among the selected strains.
Growth curve measurements
Our proteogenomic analysis identified two T7SSb-dependent effectors designated EsxA1 and EsxA2 and one T7SS target, TIGR04197, although it was not found in the culture supernatant. Therefore, we further generated ΔesxA1, ΔesxA2, ΔesxA1ΔesxA2 and ΔtigR strains in addition to ΔessC for further analysis. Bacterial cultures were prepared in BHI for 18 h and then diluted to OD600 = 0.1. Ten technical replicates of diluted culture for each strain were added to the 96-well plate and monitored for growth by measuring OD600 every 10 min for 10 h using the Varioskan LUX Multimode Microplate Reader (Thermo Scientific, USA) at 37 °C while shaking at 600 rpm. Growth rates were estimated from the slopes (log phase) obtained by fitting parametric models to the resulting data using GraphPad Prism version 8.4.3. This experiment was also done in LB and DSM cultures and done in three biological replicates.
Determination of sporulation
The sporulation process observed under aerobic conditions in DSM has been shown to induce stronger sporulation capabilities in B. cereus [36]. B. cereus ATCC14579 and its five mutants were grown at 37 °C in LB for 18 h and then diluted to OD600 = 0.1 in 10 ml DSM. The DSM culture was incubated under aerobic conditions at 30 °C overnight with shaking at 200 rpm and then upscaled to 100 ml for up to 166 h until the fraction of spores/vegetative cells reached a maximum level [36]. One milliliter of aliquots of each sample was obtained every 12 h for measuring the OD and every 24 h for establishing the colony forming units (CFU) per ml. Serial dilutions of 100 µl culture and 900 µl of 1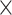 phosphate buffered saline up to 106 dilutions were made. From the 106 dilutions, an aliquot of 500 µl was heat-treated at 80 °C for 10 min to kill vegetative cells in the culture and then immediately put on ice [37, 38] while the other 500 µl remained untreated. Four 20 µl replicates of heat-treated and untreated aliquots were plated on LB and then incubated at 28 °C for 14 h to enumerate the CFUs. Complementation experiments were done in DSM supplemented with 10 µg/ml tetracycline. The mean CFU and sporulation ratios were obtained by CFU/ml = mean CFU ÷ (ml plated × dilution factor) and sporulation ratio = heat-treated mean CFU/ml ÷ untreated mean CFU/ml, respectively. Part of the undiluted daily aliquots were stained using the classical Schaeffer-Fulton staining method [39]. The slides were viewed by phase-contrast microscopy (×100, oil immersion; Nikon ELIPSE Ti, Nikon, Japan) with at least three visual fields examined to calculate the sporulation rate and images taken using a Nikon COOLPIX P310 camera. Sporulation rate = [mean spore count ÷ (mean spore count + mean vegetative cell count)] × 100%.
phosphate buffered saline up to 106 dilutions were made. From the 106 dilutions, an aliquot of 500 µl was heat-treated at 80 °C for 10 min to kill vegetative cells in the culture and then immediately put on ice [37, 38] while the other 500 µl remained untreated. Four 20 µl replicates of heat-treated and untreated aliquots were plated on LB and then incubated at 28 °C for 14 h to enumerate the CFUs. Complementation experiments were done in DSM supplemented with 10 µg/ml tetracycline. The mean CFU and sporulation ratios were obtained by CFU/ml = mean CFU ÷ (ml plated × dilution factor) and sporulation ratio = heat-treated mean CFU/ml ÷ untreated mean CFU/ml, respectively. Part of the undiluted daily aliquots were stained using the classical Schaeffer-Fulton staining method [39]. The slides were viewed by phase-contrast microscopy (×100, oil immersion; Nikon ELIPSE Ti, Nikon, Japan) with at least three visual fields examined to calculate the sporulation rate and images taken using a Nikon COOLPIX P310 camera. Sporulation rate = [mean spore count ÷ (mean spore count + mean vegetative cell count)] × 100%.
Statistical analyses
The statistical tests were performed using GraphPad Prism 8.4.3, as indicated in the figure legends, with P values < 0.05 considered significant. An ordinary unmatched one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test was applied for OD computations, and CFU counts for the same time point. A matched two-way ANOVA was used for growth kinetics, daily OD, and sporulation ratio. All experiments were conducted with at least three biological and four technical replicates.
Results
Organization and characteristics of T7SSb gene cluster in B. cereus
Gene neighborhood analysis identified candidates for the T7SSb locus in B. cereus ATCC14579 (Fig. 1). In addition, the S. aureus USA300 [12, 16] and B. subtilis 168 T7SS [20] nucleotide sequence homology analysis led to the identification of the genetic organization of T7SSb in B. cereus with notable differences. B. cereus ATCC14579 possesses two copies of the putative WXG100 protein-encoding genes locus tagged BC_RS04610 and BC_RS10395. BC_RS04610 is located > 1100 kb upstream of BC_RS10395, and the latter is immediately upstream of the T7SS core machinery genes. Immediately downstream of BC_RS04610 is the BC_RS04615, encoding a putative T7SS effector of the TIGR04197 protein family. The TIGR04197 protein sequence has no known signal peptide, interactions, or function. However, our analysis did not find TIGR04197 in the B. cereus secretome, nonetheless, we included it in the preceding experiments. Additional analysis, which included other B. cereus strains, showed that the T7SSb gene cluster is conserved up to 5’ two-thirds of the essC encoding the ATPase domain indispensable for secretion of T7SSb-dependent effectors. One-third on the 3’ end of essC and the ensuing genes within the T7SSb cluster showed variation among the less virulent B. cereus ATCC14579 and B. cereus BC33. The clinical isolates, B. cereus ST1420 and B. cereus B4264, possess a second TIGR04197 protein and LXG toxin genes within the cluster. In contrast, the variable region in S. aureus encodes other T7SSb effectors, EsxB, EsxC, EsxD, and LXG toxin, including immunity proteins (Fig. 1). LXG toxins are diverse and polymorphic, with N-terminal LXG delivery domains and diverse C-terminal toxin domains that mediate intercellular competition in biofilms via the T7SS in S. aureus and B. subtilis [40, 41].
Identification of T7SSb-dependent effectors
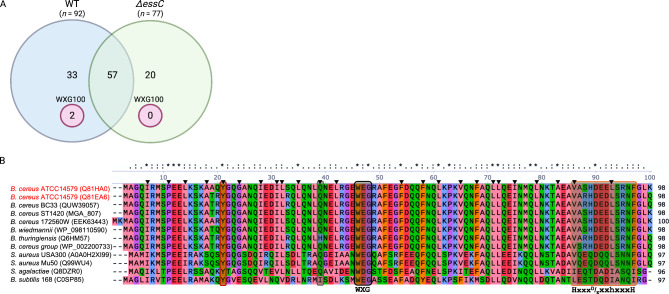
To identify the B. cereus T7SSb-dependent effector proteins, first, we generated the ΔessC mutant using B. cereus ATCC14579. The mid-log phase M9 culture of B. cereus ATCC14579 (wild type) and its ΔessC mutant were utilized in the secretome analyses to identify the T7SSb-dependent effectors. From the LC-MS/MS raw data, the search algorithm in Proteome Discoverer identified 92 proteins in the wild type (WT) and 77 proteins in the ΔessC mutant on average for three independent experiments. To increase confidence in the identifications, we limited our analysis to those proteins identified by two or more peptides. Of the 92 proteins in the secretome of wild type, only two were annotated as ESAT-6-like [Bacillus cereus] (Table S1). Motif search analysis of the secretomes in GenomeNet confirmed the two proteins designated Q81HA0 and Q81EA6 were related to T7SS. The two ESAT-6-like proteins had 94% sequence identity and were found in the WT secretome, but none in ΔessC (Fig. 2A). Proteins that did not possess known motifs related to the T7SS were excluded from further analyses. NCBI BLASTp search filtered for species diversity showed that the results comprised only firmicutes in the top 10 hits. CLUSTAL O multiple sequence alignments showed that they possessed a conserved centrally located WXG motif and C-terminal WXG100 secretion signal, HxxxD/ExxhxxxH (Fig. 2B). In addition, they had non-hydrophobic inter-dimer interacting residues at positions 21 and 39, confirming that the two effector proteins and those in the NCBI BLASTp results belong to the monocistronic expressed sagEsxA-like subfamily of the WXG100 protein superfamily [42, 43]. The proteins Q81HA0 and Q81EA6 were henceforth renamed EsxA1 and EsxA2, respectively. We confirmed that Q81HA0 and Q81EA6 were encoded in genes BC_RS04610 and BC_RS10395, respectively.
Fig. 2.
WXG100 protein, sagEsxA-like, is conserved across T7SSb loci. A. Mid-log phase secretome analysis for B cereus ATCC14579 and its ΔessC mutant. Two WXG100 proteins were found in the WT and none in ΔessC. B. CLUSTAL O alignments of the two WXG100 protein sequences, Q81HA0 (EsxA1) and Q81EA6 (EsxA2), found in the WT secretome and other firmicutes species. The alignment shows the centrally located WXG motif and a conserved C-terminal secretion signal with the HxxxD/ExxhxxxH. In this sequence, ‘H’ stands for highly conserved hydrophobic and ‘h’ for less conserved hydrophobic residues, ‘x’ for any amino acid, and ‘D/E’ for either aspartic or glutamic acids, respectively. The four-helix bundle requires predominantly hydrophobic residues at a helix turn consisting of the heptad helix repeat shown by ▼ on the aligned residues. All residues involved in the inter-dimer interactions are hydrophobic except two residues, position 21 and 39, which are unique to the sagEsxA-like subfamily
Colony characteristics and growth patterns under normal conditions
The genes BC_RS04610, BC_RS10395, and BC_RS04615 were renamed esxA1, esxA2, and tigR, respectively. We successfully generated B. cereus ATCC14579 full gene deletions in esxA1, esxA2, esxA1esxA2, and tigR in addition to the ΔessC mutant for further analyses. Some studies on the functional diversity of T7SS-dependent substrates have insinuated that secreted effectors may have multiple roles besides being just secreted virulence effectors [13, 44, 45]. Therefore, we screened the T7SSb mutant strains against the WT for morphotype changes and growth patterns’ aberrations. When checked for colony characteristics, the WT and the mutant strains had similar colony size and morphology in BHI agar, and when plated on blood agar, they were all surrounded by a halo of hemolysis (Figs. S2A and S2B). Growth on cereus selective agar base supplemented with 100 units/ml polymyxin B and 5% egg-yolk emulsion (Merk, Germany), all strains induced egg-yolk precipitation with purple colonies and surrounding media (Fig. S2C). Simple linear regression of the log phase growth in minutes for all strains (n = 10) in BHI (60–150), LB (70–150), and DSM (60–170) did not show any significant differences regardless of the media used. Since the slopes were not significantly different from each other, their pooled values of the log phases were 0.0058 (R2 = 0.995), 0.0082 (R2 = 0.994), and 0.0067 (R2 = 0.988), in BHI, LB, and DSM, respectively. Thus, the gene deletions in this study did not alter the growth rate, suggesting T7SSb does not play a role in the growth of B. cereus. The growth curves and their respective simple linear regression log phase slopes are shown in (Fig. S3).
Growth under sporulation conditions was impaired in ΔesxA1ΔesxA2
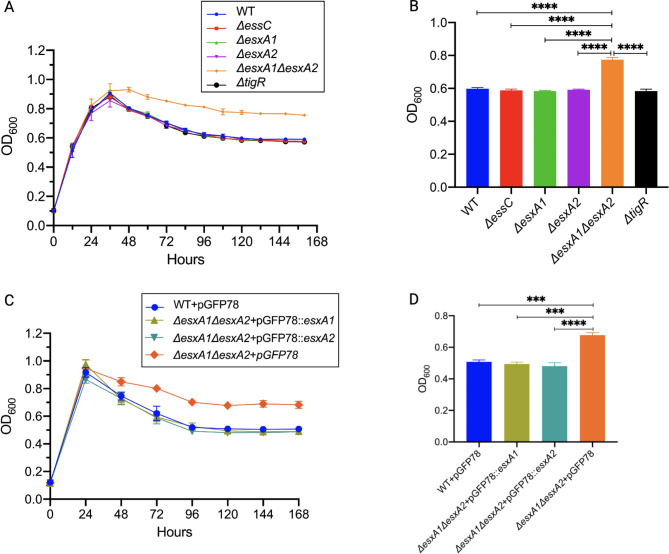
Under normal growth conditions, the T7SSb mutants and the WT were indistinguishable; therefore, we next checked growth under nutritional stress conditions that stimulated sporulation. Growth in DSM was monitored by measuring OD600 every 12 h until a stable peak measurement. Figure 3A shows an exponential increase in OD in the first 36 h, followed by a sharp decrease in all strains except ΔesxA1ΔesxA2 mutant, which exhibited a slow decline before becoming nearly constant after 120 h. At 120 h, ΔesxA1ΔesxA2 had a significantly higher OD than other strains (Fig. 3B). The decrease in OD represented the sporulation process as mother cells lyse, releasing mature spores and shifting from the denser vegetative cells to smaller and lighter spores. Complementation of esxA1esxA2 mutation with either esxA1 or esxA2 resulted in ODs comparable to the WT (Fig. 3C and D). The significantly higher OD in the ΔesxA1ΔesxA2 mutant could be due to delayed autolysis by the mother cell required to release mature spores. In assessing this hypothesis, we recorded the spore ratio from CFUs and observed the sporulation rate microscopically recorded every 24 h. The highest CFU/ml for the untreated and heat-treated cultures was observed after 120 h, at which their ratio was maximal for all the strains. Extended cultures did not yield different results, thus, the CFU data presented here were measured after 120 h of culture, which coincided with the stable OD (Fig. 4A). The ΔesxA1ΔesxA2 mutant had a significantly lower spore ratio (~ 0.72), signifying fewer viable spores per ml. In contrast, ΔesxA1, ΔesxA2, ΔessC and ΔtigR mutants had spore ratio results comparable to the WT at all time points, peaking at approximately 0.96 (Fig. 4A). There was a significant reduction in the mean CFU/ml for the ΔesxA1ΔesxA2 mutant when heat treated, indicating the presence of vegetative cells after 120 h (Fig. 4B and C). Such a reduction was not observed in the WT and other mutants. When complemented, maximum sporulation rates (97%) were restored in both ΔesxA1ΔesxA2 + pGFP78::esxA1 and ΔesxA1ΔesxA2 + pGFP78::esxA2 strains, demonstrating that esxA1 and esxA2 worked cooperatively and either was sufficient to restore the sporulation phenotype (Fig. 4D – F). Our findings indicated that EsxA was essential for optimum spore formation in B. cereus ATCC14579.
Fig. 3.
Optical density measurement during culture in DSM at 30 °C. A. Optical density throughout the growth of B. cereus ATCC14579 and its mutants in DSM. B. Optical density measured after 120 h culture. C. Time-course and (D) at 120 h optical density for complemented strains. The results represent means ± SEM of 3 independent experiments. Statistical significance was calculated by Ordinary one-way ANOVA with Dunnett’s test, **P = 0.002; ***P = 0.0003***P = 0.0002; ****P < 0.0001
Fig. 4.
Sporulation rates and spore viability of experiment strains cultured in DSM at 30 °C. A. Time course sporulation rates were calculated as the ratio of CFUs/ml for heat-treated and untreated WT and its mutants. The mean CFU/ml for untreated (B) and heat-treated (C) cultures obtained after 120 h of culture. D. Time course sporulation rates for complemented strains and their mean CFUs/ml for the untreated (E) and heat-treated cultures (F). Statistical significance was calculated using Ordinary one-way ANOVA with Dunnett’s multiple comparisons test, *P < 0.0382; **P < 0.0093; ***P < 0.006 for three independent experiments
EsxA is essential for optimum sporulation in B. cereus ATCC14579
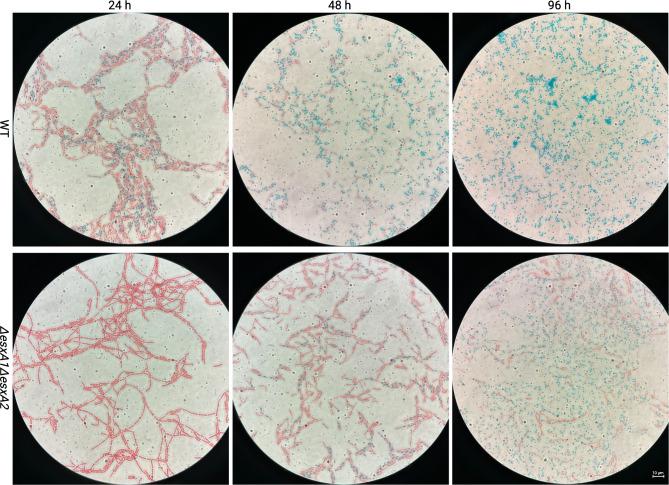
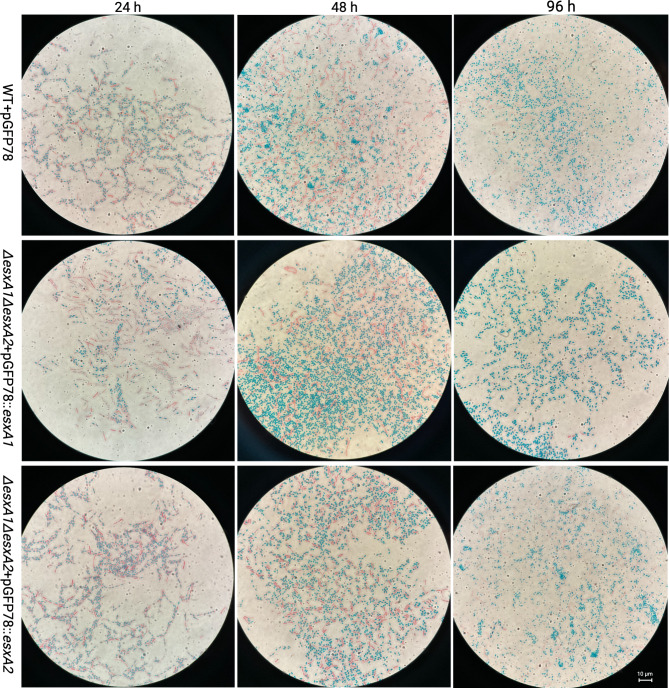
We found that the ΔesxA1ΔesxA2 mutant had a significantly higher OD but a significantly lower CFU/ml, fewer spores, and a lower sporulation ratio. We conducted microscopy of the Schaeffer-Fulton method-stained cultures to evaluate our delayed mother cell autolysis hypothesis. Phase contrast microscopy showed early evidence of sporulation in the majority (> 80%) of the WT cells generating endospores within 24 h compared to ΔesxA1ΔesxA2 with less than 10%. The ΔesxA1ΔesxA2 mutant endospores poorly took up malachite green stain at this time, suggesting fewer endospores (Fig. 5). After culturing for 48 h, WT released a mass of mature spores, but ΔesxA1ΔesxA2 had a few mature spores, and almost 55% of the cells had endospores. Further, the ΔesxA1ΔesxA2 mutant had vegetative cells beyond 96 h, which had maintained the typical chaining phenotype. The presence of vegetative cells accounted for the significantly higher OD and a reduced sporulation rate, whose peak was 72%. In contrast, at 96 h, the WT sporulation rate almost reached 100%. These findings ruled out our hypothesis, suggesting that delayed autolysis of mother cells with mature endospores caused a significantly higher OD and lower spore ratio, but rather, due to an inefficient sporulation process altogether. Again, complementation of ΔesxA1ΔesxA2 with either esxA1 or esxA2 reversed the sporulation deficiency to levels of the WT (Fig. 6). Therefore, we concluded that the two EsxA proteins work cooperatively and play a significant role in the optimal sporulation of B. cereus ATCC14579.
Fig. 5.
The spore staining results of B. cereus ATCC14579 and ΔesxA1ΔesxA2. When stained with malachite green and safranin O, matured spores and endospores are stained blue, and vegetative cells are stained red. Within 24 h, more than 80% of the WT cells had formed endospores (blue cytoplasm), but less than 10% in ΔesxA1ΔesxA2 cells, which are immature (forespores) and poorly take up the malachite green stain. After 48 h, the WT underwent 66% sporulation on average, while the mutant had approximately 45% endospores and 20% mature spores. Beyond 96 h, the WT had almost all cells sporulated, but ΔesxA1ΔesxA2 still had vegetative cells (28%) occurring in chains
Fig. 6.
The spore staining results of complementary strains. Complementation of ΔesxA1ΔesxA2 with either pGFP78::esxA1 or pGFP78::esxA2 restored sporulation rates to levels of the WT. Spores and endospores are stained blue, while vegetative cells red. The WT was also complemented with pGFP78 to allow growth of all complemented strains under similar conditions in 10 µg/ml tetracycline
Discussion
The T7SS is characterized by an ATPase-driven secretion of approximately 100-residue helical proteins that contain a central WXG motif, with EsxA being the first identified T7SS substrate [46]. It is the major effector of the T7SS secretion system, essential for the virulence of pathogenic Gram-positive bacteria, such as M. tuberculosis and S. aureus. EsxA is required for the synthesis of EsxB and vise-versa, interacts with multiple cellular proteins, and stimulates several signal pathways [47–50]. This greatly increases the complexity of dissecting the precise roles of EsxA. To date, no study has focused on the functional roles of EsxA nor characterized the T7SSb in B. cereus.
In this study, we focused on establishing the repertoire of the T7SSb in B. cereus ATCC14579 and investigating the intrinsic roles of the T7SSb-dependent effector proteins. Through LC-MS/MS concatenated to a proteogenomic pipeline, we identified two EsxA proteins with 94% sequence identity in the wild-type culture supernatant and none in the ΔessC mutant. Protein sequence alignment showed that this pair belonged to the sagEsxA-like subfamily of the WXG100 superfamily. WXG100 proteins form dimeric complexes within the cytoplasm before secretion to increase solubility, and the driving force for the interdimer interactions is principally hydrophobic [42, 51]. However, a pair of conserved and hydrophilic mutations among the proteins of the sagEsxA-like subfamily allows for interdimer hydrogen bonding. As a result, this conserved pair of mutations is seen as a fingerprint of homodimeric WXG100 proteins, which is in tandem with our finding of two nearly identical EsxA proteins (Fig. 2B). In contrast, ESAT-6-like and EsxB found in S. aureus and M. tuberculosis form heterodimers that only possess hydrophobic residues for interdimer interaction [42]. Our genome analysis did not find an EsxB candidate, similarly, B. subtilis T7SS lacks the EsxB homolog (Fig. 1) [19, 20]. Among Bacillus species, EsxB has only been identified in B. anthracis, but it lacks EsxA [21]. In addition, some members of the Firmicutes group have the atypical WXG100-like proteins, EsxC and EsxD, encoded on the same operon or within the T7SS gene cluster [16]. However, in B. cereus, a TIGR04197 protein family gene was found downstream of the orphaned esxA1, sharing the same operon (Fig. 1). The protein sequence has no known signal peptide or interactions but based on the curated Hidden Markov Model-based and BLAST-based protein families, members of the TIGR04197 protein family are similar in length and sequence. Phylogenetic profiling shows that members of this family are similarly restricted to species with T7SS, making this family a related set of T7SS effectors.
Homology analysis among B. cereus strains showed that the T7SSb gene cluster is conserved from the 5’ end up to the proximal two-thirds of the essC. One-third on the 3’ end of essC and the ensuing genes within the T7SSb cluster showed variation among the strains. This could suggest that B. cereus strains have different essC alleles, with each allele followed by a distinct set of genes. The less virulent B. cereus ATCC14579 and B. cereus BC33 did not possess virulence-encoding genes in the variable region. In contrast, the clinical isolates, B. cereus ST1420 and B. cereus B4264, possess a second TIGR04197 protein and LXG toxin genes (Fig. 1). B. cereus ST1420 was a major sequence type associated with nosocomial infections and bacteremia in Japan [2], while B. cereus B4264 was isolated from a case of fatal pneumonia in a male patient cultured from the blood and the pleural fluid in the USA [52]. The variable 3’ end of the essC encodes the C-terminal ATPase domain to which the substrate binds before secretion. The EssC variants among Firmicutes are associated with unique effector repertoires, it is likely that a given EssC may only export substrates encoded by their cognate subtype. Studies on T7SSb in S. aureus and Listeria monocytogenes strains have identified multiple essC alleles defined by downstream substrates. S. aureus and L. monocytogenes strains have been grouped into four and seven T7SSb variants, respectively [16, 53, 54]. S. aureus strains with essC1 are clinical isolates, possessing esxA, esxB, esxC, esxD, and an LXG toxin gene followed by immunity genes, while the less pathogenic strains with essC2 and essC3, the esxD is replaced by tigR [16, 53]. TIGR04197 could play a significant role, yet to be known, in the T7SSb function. Our findings, and those of others, show that the T7SSb is very diverse between and within species, a reflection of virulence and implications for varied functions among the Firmicutes group.
Recent studies on T7SS continue to focus on its involvement in host-immune interactions and niche adaptations, which are diverse given the inter and intraspecies variations in the repertoire. We also need to start unraveling the intrinsic roles of the T7SS, which could provide insights into the pathways they are involved in and serve as new targets for antimicrobials in the wake of emerging drug-resistant bacteria [55, 56]. Here, we screened for alterations in the phenotypic characteristics that gene deletions in the secretion system might cause in B. cereus ATCC14579. We found colony size and morphology, hemolytic and proteolytic activities, and growth kinetics similar to the wild type. When tested for sporulation in DSM, we found that the strain lacking both esxA genes, ΔesxA1ΔesxA2, had a delayed and significantly reduced ability to form spores. It was evident from the spore ratio that ΔesxA1ΔesxA2 had significantly fewer viable spores throughout the culture period compared to the WT and other mutants (Fig. 4). Generally, a higher bacterial culture optical density is assumed to translate to a higher CFU count, however, our findings for the ΔesxA1ΔesxA2 defied that notion. Phase contrast microscopy showed no evident aberrations in cell and spore morphologies, only a delayed and reduced sporulation in ΔesxA1ΔesxA2, which agreed with the CFU results. Overall, the ΔesxA1ΔesxA2 culture still had vegetative cells reaching 28% on average of any microscopic field viewed (Fig. 5). When complemented with either esxA1 or esxA2, the mutant fully recovered the spore formation ability (Fig. 6), suggesting the cooperative function of the EsxA1 and EsxA2 and their involvement in sporulation of B. cereus. In contrast, the ΔessC mutant, in which we found that EsxA was not secreted, had similar sporulation results as the WT. This indicated that EsxA did not affect sporulation after being secreted, but its expression within the cell was necessary to maintain optimum sporulation. Thus, ΔesxA1, ΔesxA2, and ΔtigR sporulation phenotypes were unaffected. These findings confirmed that EsxA is not only a secreted effector protein but has additional roles within the milieu and, in this case, sporulation. Other studies have also indicated that T7SS-dependent effectors have other roles within the bacterium. Fyans et al. [57] reported that the ESX/type VII secretion system modulated development but not virulence in Streptomyces scabies, a plant pathogen. Specifically, the EsxA and EsxB mutants had abnormal spore chains and exhibited resistance to lysis by the Streptomyces-specific phage ϕC31. Similarly, deletion of the EsxAB operon in Streptomyces coelicolor resulted in irregular-sized pre-spore compartments with corresponding aberrant DNA contents. The EsxAB heterodimer was proposed to control the coordination of cell division with the segregation of nucleoids, most probably through interaction with other factors that regulate nucleoid condensation [58]. However, in B. cereus, growth, size, and morphology of the colonies and cells of the ΔesxA1ΔesxA2 mutant were unaffected. The T7SS-mediated phenotypes seemingly depend on T7SS subtype-specific regulation and secreted effectors, a pandora’s box yet to be opened. Thus, T7SS substrates possess diverse species-specific cellular functions and can modulate interactions between bacteria and eukaryotic cells [44, 47, 49].
One possible mechanism of how EsxA plays an important role in the sporulation in B. cereus is that EsxA may interact with SpoIIIE and enhance its functionality. The protein-protein interaction analysis using STRING [59] (https://string-db.org) showed that YukE, an EsxA from B. subtilis, may interact with SpoIIIE (Fig. S4A). SpoIIIE is a DNA translocase that drives up to 70% of the chromosome across the forespore septum before endospore formation [60, 61]. Since B. cereus EsxA proteins were unable to be analyzed in the STRING database, and the sporulation pathway is well understood in B. subtilis [62–65], the prediction data was obtained using B. subtilis protein information. Although the amino acid sequences of EsxA and YukE have low homology, the superimposition of their predicted structures was nearly perfect (Fig.S4B). Further, the SpoIIIE proteins from B. cereus and B. subtilis were predicted to possess a high structural homology (Fig. S4C), therefore, we speculate that EsxA-SpoIIIE interaction may occur, and it impact sporulation in B. cereus. In addition, we also found that the ATPase domain of B. cereus SpoIIIE had a high structural similarity with the ATPase domain of EssC protein, as Mietrach et al. revealed previously [66]. Just like EsxB binds to EssC, causing the multimerization of EssC to form the hexameric secretion pore [24, 67], SpoIIIE also forms a hexameric pore to pass DNA [13, 23]. Hence, in the absence of EsxB in B. cereus ATCC14679, we conjecture that EsxA may promote EssC and SpoIIIE multimerization. Previously, it was observed in S. coelicolor that the loss of function of the EsxAB heterodimer impacted the coordination of cell division and the segregation of nucleoids. But even multiple gene mutations involved in the coordination of nucleoid condensation and segregation, including ftsK, also resulted in similar septation aberrations, implying it depends on several overlapping and partially redundant functions [58]. Similarly, in B. cereus ATCC14579, loss of EsxA1 and EsxA2 may be impacting the sporulation function of SpoIIIE, an FtsK protein. Thus, without EsxA in the ΔesxA1ΔesxA2 mutant, SpoIIIE translocation of chromosomal DNA may have been inefficient, resulting in delayed sporulation and fewer mature spores in B. cereus ATCC14579. On the other hand, EsxA might have a completely different novel mechanism affecting sporulation, which regulates the function or transcription of sporulation-related genes. The detailed molecular mechanisms of how EsxA regulates B. cereus sporulation and its biological significance in the B. cereus lifecycles, including the virulence, need to be addressed in future studies.
Conclusion
B. cereus T7SSb possesses a pair of nearly identical effector proteins named EsxA belonging to the sagEsxA-like subfamily of the WXG100 protein superfamily. Single gene mutations in the T7SSb effectors and core machinery did not alter the growth characteristics of B. cereus ATCC14579. However, deletion of the two esxA genes resulted in a significantly delayed sporulation and fewer viable spores/ml in a sporulation-inducing culture. Additionally, complementation of the ΔesxA1ΔesxA2 mutant with a single esxA gene was sufficient to recover maximum sporulation ability. Altogether, these results consistently showed that EsxA is essential for spore formation in B. cereus ATCC14579, a novel finding for a T7SSb-dependent effector molecule.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the Open Facility, Global Facility Center, Creative Research Institution, Hokkaido University for allowing us to conduct the analysis of peptide mixture using the HPLC and Hybrid Ion Trap-Orbitrap Mass Spectrometer, which greatly assisted the research. We are grateful to Yunrong Chai and Yinghao He (Northeastern University, Boston) for generously providing the pGFP78 DNA.
Author contributions
H.K.K. and H.H. conceptualized the study. H.K.K., A.P. and Y.F. designed the experiments. H.K.K., M.Su. and Y.F. carried out the experiments. H.K.K., M.Sh., J.Y.C., T.Z. and T.K. contributed to data analysis and interpretation of results. H.K.K., A.P., and M.Sh. drafted the manuscript, while Y.F., M.M., B.M.H., T.K. and A.P. reviewed and edited it. H.H. and A.P. supervised the study. All authors read and approved the final manuscript.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) in Japan or Japan Society for the Promotion of Science under Grants-in-Aid for Scientific Research (KAKENHI) to H.H. (Grant No.17H01679 and 18K19436) and A.P. (Grant No. 19K16653 and 21K15430), World-leading Innovative and Smart Education (WISE) Grant-in-Aid for Graduate Students to H.K.K., and the Japan International Cooperation Agency (JICA) scholarship for Advanced Training Program for Fostering Global Leaders on Infectious Disease Control to Build Resilience against Public Health Emergencies to H.K.K.
Data availability
The B. cereus (ATCC14579, BC33, B4262, and ST1420), S. aureus USA300, and B. subtilis 168 genome datasets used in the analysis of the type VIIb secretion system were downloaded from NCBI Genome under the GenBank file numbers NC_004722.1, CP072774.1, CP001176.1, AP022975.1, CP092052.1, and AL009126.3 respectively.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Acosta Pedemonte NB, Rocchetti NS, Villalba J, Lerman Tenenbaum D, Settecase CJ, Bagilet DH, et al. Bacillus cereus bacteremia in a patient with an abdominal stab wound. Rev Argent Microbiol. 2020;52(2):115–7. [DOI] [PubMed] [Google Scholar]
- 2.Akamatsu R, Suzuki M, Okinaka K, Sasahara T, Yamane K, Suzuki S, et al. Novel sequence type in Bacillus cereus strains associated with nosocomial infections and bacteremia, Japan. Emerg Infect Dis. 2019;25(5):883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klee SR, Ozel M, Appel B, Boesch C, Ellerbrok H, Jacob D, et al. Characterization of Bacillus anthracis-like bacteria isolated from wild great apes from Cote d’Ivoire and Cameroon. J Bacteriol. 2006;188(15):5333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enosi Tuipulotu D, Mathur A, Ngo C, Man SM. Bacillus cereus: epidemiology, virulence factors, and host–Pathogen interactions. Trends Microbiol. 2021;29(5):458–71. [DOI] [PubMed] [Google Scholar]
- 5.Abdallah AM, van Gey NC, Champion PAD, Cox J, Luirink J, Vandenbroucke-Grauls CMJE, et al. Type VII secretion–mycobacteria show the way. Nat Rev Microbiol. 2007;5(11):883–91. [DOI] [PubMed] [Google Scholar]
- 6.Bunduc CM, Bitter W, Houben ENG. Structure and function of the mycobacterial type VII Secretion systems. Annu Rev Microbiol. 2020;74(1):315–35. [DOI] [PubMed] [Google Scholar]
- 7.Kneuper H, Cao ZP, Twomey KB, Zoltner M, Jäger F, Cargill JS, et al. Heterogeneity in ess transcriptional organization and variable contribution of the Ess/Type VII protein secretion system to virulence across closely related Staphylococcus aureus strains. Mol Microbiol. 2014;93(5):928–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spencer BL, Tak U, Mendonça JC, Nagao PE, Niederweis M, Doran KS. A type VII secretion system in Group B Streptococcus mediates cytotoxicity and virulence. PLoS Pathog. 2021;17(12):e1010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol. 2002;46(3):709–17. [DOI] [PubMed] [Google Scholar]
- 10.Smith J, Manoranjan J, Pan M, Bohsali A, Xu J, Liu J, et al. Evidence for pore formation in host cell membranes by ESX-1-Secreted ESAT-6 and its role in Mycobacterium marinum escape from the Vacuole. Infect Immun. 2008;76(12):5478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tak U, Dokland T, Niederweis M. Pore-forming esx proteins mediate toxin secretion by Mycobacterium tuberculosis. Nat Commun. 2021;12(1):394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol. 2016;2(1):16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan H, Mohamed AMT, Grainge I, Rodrigues CDA. FtsK and SpoIIIE, coordinators of chromosome segregation and envelope remodeling in bacteria. Trends Microbiol. 2022;30(5):480–94. [DOI] [PubMed] [Google Scholar]
- 14.Callahan B, Nguyen K, Collins A, Valdes K, Caplow M, Crossman DK, et al. Conservation of structure and protein-protein interactions mediated by the secreted mycobacterial proteins EsxA, EsxB, and EspA. J Bacteriol. 2010;192(1):326–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champion PAD, Stanley SA, Champion MM, Brown EJ, Cox JS. C-Terminal Signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Sci (1979). 2006;313(5793):1632–6. [DOI] [PubMed] [Google Scholar]
- 16.Warne B, Harkins CP, Harris SR, Vatsiou A, Stanley-Wall N, Parkhill J, et al. The Ess/Type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genomics. 2016;17(1):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bobrovskyy M, Chen X, Missiakas D. The type 7b secretion system of S. Aureus and its role in colonization and systemic infection. Infect Immun. 2023;91(5). [DOI] [PMC free article] [PubMed]
- 18.Klein TA, Grebenc DW, Shah PY, McArthur OD, Dickson BH, Surette MG et al. Dual targeting factors are required for LXG Toxin Export by the bacterial type VIIb secretion system. mBio. 2022;13(5). [DOI] [PMC free article] [PubMed]
- 19.Tassinari M, Doan T, Bellinzoni M, Chabalier M, Ben-Assaya M, Martinez M et al. The antibacterial type VII secretion system of Bacillus subtilis: structure and interactions of the pseudokinase YukC/EssB. mBio. 2022;13(5). [DOI] [PMC free article] [PubMed]
- 20.Huppert LA, Ramsdell TL, Chase MR, Sarracino DA, Fortune SM, Burton BM. The ESX System in Bacillus subtilis mediates protein secretion. PLoS ONE. 2014;9(5):e96267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garufi G, Butler E, Missiakas D. ESAT-6-Like protein secretion in Bacillus anthracis. J Bacteriol. 2008;190(21):7004–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleming TC, Shin JY, Lee SH, Becker E, Huang KC, Bustamante C, et al. Dynamic SpoIIIE assembly mediates septal membrane fission during Bacillus subtilis sporulation. Genes Dev. 2010;24(11):1160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiche JB, Cattoni DI, Diekmann N, Langerak JM, Clerte C, Royer CA, et al. Recruitment, Assembly, and Molecular Architecture of the SpoIIIE DNA pump revealed by Superresolution Microscopy. PLoS Biol. 2013;11(5):e1001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberg OS, Dovala D, Li X, Connolly L, Bendebury A, Finer-Moore J, et al. Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell. 2015;161(3):501–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholson WL, Setlow P, Harwood C, Cutting SM. Molecular biological methods for Bacillus. New York, USA: John Willey; 1990. pp. 391–450. [Google Scholar]
- 26.Plaut RD, Stibitz S. Improvements to a markerless allelic exchange system for Bacillus anthracis. PLoS ONE. 2015;10(12):e0142758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao T, Foulston L, Chai Y, Wang Q, Losick R. Alternative modes of biofilm formation by plant-associated Bacillus cereus. Microbiologyopen. 2015;4(3):452–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature. 2003;423(6935):87–91. [DOI] [PubMed] [Google Scholar]
- 29.Simon R, Priefer U, Pühler A. A Broad Host Range Mobilization System for in Vivo Genetic Engineering: Transposon Mutagenesis in Gram negative Bacteria. Bio/Technology. 1983;1(9):784–91. [Google Scholar]
- 30.Chan QWT, Howes CG, Foster LJ. Quantitative Comparison of Caste Differences in Honeybee Hemolymph. Mol Cell Proteom. 2006;5(12):2252–62. [DOI] [PubMed] [Google Scholar]
- 31.Deng W, Yu HB, de Hoog CL, Stoynov N, Li Y, Foster LJ, et al. Quantitative proteomic analysis of type III secretome of Enteropathogenic Escherichia coli reveals an expanded Effector Repertoire for Attaching/Effacing bacterial pathogens. Cell Proteom. 2012;11(9):692–709. Molecular. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian C, Chen H, Johs A, Lu X, An J, Pierce EM, et al. Quantitative proteomic analysis of biological processes and responses of the Bacterium Desulfovibrio desulfuricans ND132 upon deletion of its Mercury methylation genes. Proteomics. 2018;18(17):1700479. [DOI] [PubMed] [Google Scholar]
- 33.Kanehisa M. Linking databases and organisms: GenomeNet resources in Japan. Trends Biochem Sci. 1997;22(11):442–4. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. [DOI] [PubMed] [Google Scholar]
- 35.Guy L, Roat Kultima J, Andersson SGE. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26(18):2334–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Jin J, Hu H, Deveau IF, Foley SL, Chen H. Optimization of sporulation and purification methods for sporicidal efficacy assessment on Bacillus spores. J Ind Microbiol Biotechnol. 2022;49(4). [DOI] [PMC free article] [PubMed]
- 37.Huang Q, Zhang Z, Liu Q, Liu F, Liu Y, Zhang J, et al. SpoVG is an important regulator of sporulation and affects biofilm formation by regulating Spo0A transcription in Bacillus cereus. BMC Microbiol. 2021;21(1):0–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholson WL, Schuerger AC. Bacillus subtilis spore survival and expression of Germination-Induced Bioluminescence after prolonged incubation under simulated Mars Atmospheric pressure and composition: implications for Planetary Protection and Lithopanspermia. Astrobiology. 2005;5(4):536–44. [DOI] [PubMed] [Google Scholar]
- 39.Oktari A, Supriatin Y, Kamal M, Syafrullah H. The bacterial endospore stain on Schaeffer Fulton using variation of Methylene Blue Solution. J Phys Conf Ser. 2017;812:012066. [Google Scholar]
- 40.Ulhuq FR, Gomes MC, Duggan GM, Guo M, Mendonca C, Buchanan G, et al. A membrane-depolarizing toxin substrate of the Staphylococcus aureus type VII secretion system mediates intraspecies competition. Proc Natl Acad Sci. 2020;117(34):20836–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi K. Diverse LXG toxin and antitoxin systems specifically mediate intraspecies competition in Bacillus subtilis biofilms. PLoS Genet. 2021;17(7):e1009682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poulsen C, Panjikar S, Holton SJ, Wilmanns M, Song YH. WXG100 protein superfamily consists of three subfamilies and exhibits an α-Helical C-Terminal conserved Residue Pattern. PLoS ONE. 2014;9(2):e89313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sysoeva TA, Zepeda-Rivera MA, Huppert LA, Burton BM. Dimer recognition and secretion by the ESX secretion system in Bacillus subtilis. Proceedings of the National Academy of Sciences. 2014;111(21):7653–8. [DOI] [PMC free article] [PubMed]
- 44.Spencer BL, Doran KS. Evolving understanding of the type VII secretion system in Gram-positive bacteria. PLoS Pathog. 2022;18(7):e1010680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flint JL, Kowalski JC, Karnati PK, Derbyshire KM. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proceedings of the National Academy of Sciences. 2004;101(34):12598–603. [DOI] [PMC free article] [PubMed]
- 46.Unnikrishnan M, Constantinidou C, Palmer T, Pallen MJ. The enigmatic esx proteins: looking Beyond Mycobacteria. Trends Microbiol. 2017;25(3):192–204. [DOI] [PubMed] [Google Scholar]
- 47.Burts ML, Williams WA, DeBord K, Missiakas DM. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci. 2005;102(4):1169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korea CG, Balsamo G, Pezzicoli A, Merakou C, Tavarini S, Bagnoli F, et al. Staphylococcal esx proteins modulate apoptosis and release of Intracellular Staphylococcus aureus during infection in epithelial cells. Infect Immun. 2014;82(10):4144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, Wang D, Jiang G, Liu W, Deng Q, Li X, et al. EsxA membrane-permeabilizing activity plays a key role in mycobacterial cytosolic translocation and virulence: effects of single-residue mutations at glutamine 5. Sci Rep. 2016;6(1):32618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bao Y, Wang L, Sun J. A small protein but with diverse roles: a review of EsxA in Mycobacterium–host Interaction. Cells. 2021;10(7):1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renshaw PS, Panagiotidou P, Whelan A, Gordon SV, Hewinson RG, Williamson RA, et al. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis Complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the Structural properties of ESAT-6, CFP-10, and the ESAT-6·CFP-10 complex. J Biol Chem. 2002;277(24):21598–603. [DOI] [PubMed] [Google Scholar]
- 52.Dodson RJ, Durkin AS, Rosovitz MJ, Rasko DA, Hoffmaster A, Ravel J et al. Genome sequence of Bacillus cereus B4264. Vol. Direct sub, NCBI. Maryland; 2008.
- 53.Bowman L, Palmer T. The type VII secretion system of Staphylococcus. Annu Rev Microbiol. 2021;75(1):471–94. [DOI] [PubMed] [Google Scholar]
- 54.Bowran K, Palmer T. Extreme genetic diversity in the type VII secretion system of Listeria monocytogenes suggests a role in bacterial antagonism. Microbiol (N Y). 2021;167(3). [DOI] [PMC free article] [PubMed]
- 55.Baron C, Coombes B. Targeting bacterial Secretion systems: benefits of disarmament in the Microcosm. Infect Disord Drug Targets. 2007;7(1):19–27. [DOI] [PubMed] [Google Scholar]
- 56.Belete TM. Novel targets to develop new antibacterial agents and novel alternatives to antibacterial agents. Hum Microb J. 2019;11:100052. [Google Scholar]
- 57.Fyans JK, Bignell D, Loria R, Toth I, Palmer T. The ESX type VII secretion system modulates development, but not virulence, of the plant pathogen Streptomyces scabies. Mol Plant Pathol. 2013;14(2):119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akpe San Roman S, Facey PD, Fernandez-Martinez L, Rodriguez C, Vallin C, Del Sol R, et al. A heterodimer of EsxA and EsxB is involved in sporulation and is secreted by a type VII secretion system in Streptomyces coelicolor. Microbiol (N Y). 2010;156(6):1719–29. [DOI] [PubMed] [Google Scholar]
- 59.Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 1995;9(11):1316–26. [DOI] [PubMed] [Google Scholar]
- 61.Wu LJ, Errington J. Bacillus subtilis spoIIIE protein required for DNA segregation during Asymmetric Cell Division. Sci (1979). 1994;264(5158):572–5. [DOI] [PubMed] [Google Scholar]
- 62.Eijlander RT, de Jong A, Krawczyk AO, Holsappel S, Kuipers OP. SporeWeb: an interactive journey through the complete sporulation cycle of Bacillus subtilis. Nucleic Acids Res. 2014;42(D1):D685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36(1):131–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riley EP, Schwarz C, Derman AI, Lopez-Garrido J. Milestones in Bacillus subtilis sporulation research. Microb Cell. 2021;8(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Omony J, de Jong A, Krawczyk AO, Eijlander RT, Kuipers OP. Dynamic sporulation gene co-expression networks for Bacillus subtilis 168 and the food-borne isolate Bacillus amyloliquefaciens: a transcriptomic model. Microb Genom. 2018;4(2). [DOI] [PMC free article] [PubMed]
- 66.Mietrach N, Damián-Aparicio D, Mielich-Süss B, Lopez D, Geibel S. Substrate Interaction with the EssC coupling protein of the type VIIb secretion system. J Bacteriol. 2020;202(7). [DOI] [PMC free article] [PubMed]
- 67.Bobrovskyy M, Oh SY, Missiakas D. Contribution of the EssC ATPase to the assembly of the type 7b secretion system in Staphylococcus aureus. J Biol Chem. 2022;298(9):102318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The B. cereus (ATCC14579, BC33, B4262, and ST1420), S. aureus USA300, and B. subtilis 168 genome datasets used in the analysis of the type VIIb secretion system were downloaded from NCBI Genome under the GenBank file numbers NC_004722.1, CP072774.1, CP001176.1, AP022975.1, CP092052.1, and AL009126.3 respectively.