Figure 1. Genome-wide CRISPR-Cas9 screens identify the GCN2 pathway as a mediator of the response of SCLC cells to WEE1 kinase inhibition.
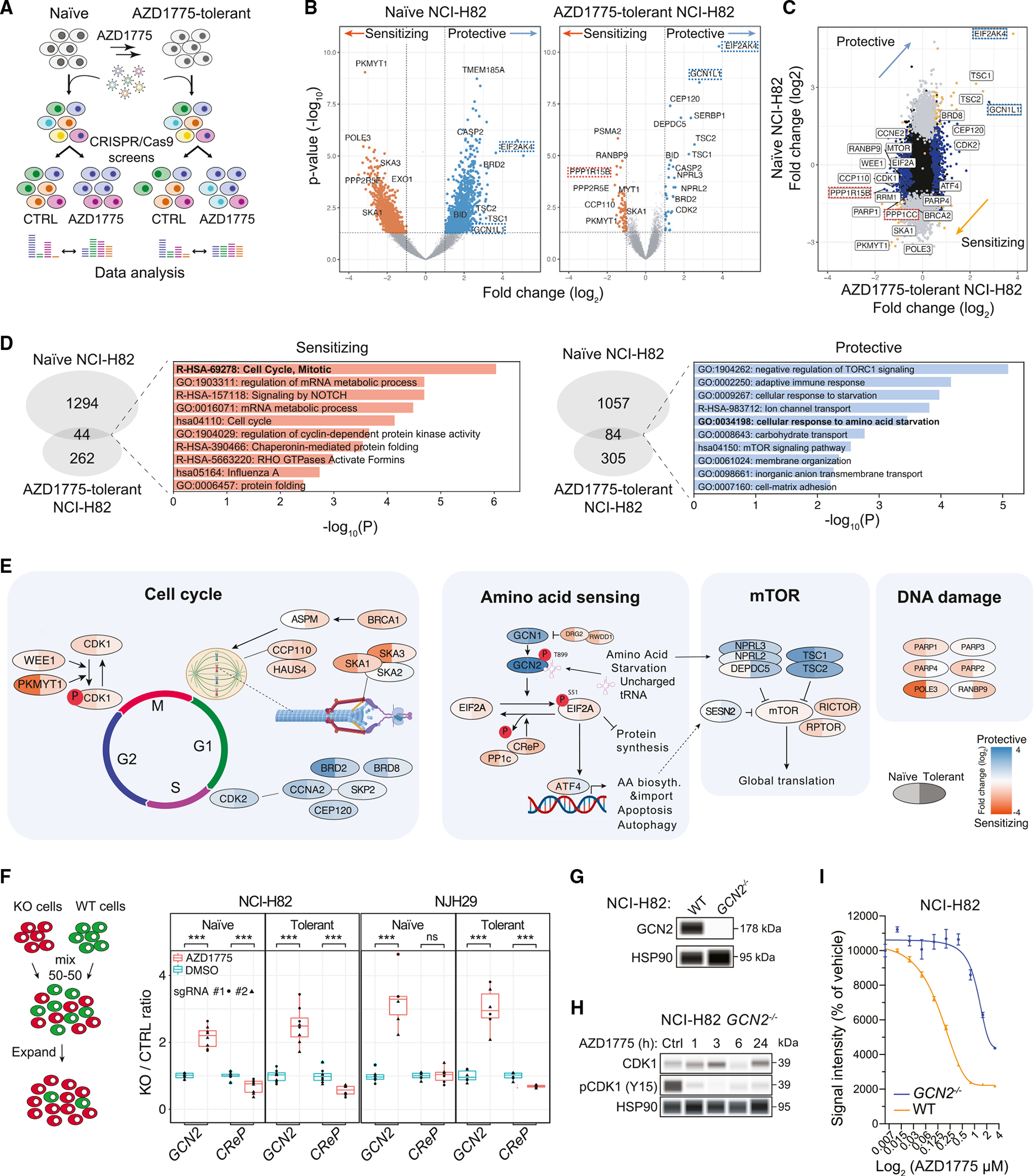
(A) CRISPR-Cas9 genome-wide screening strategy.
(B) Volcano plots from genome-wide CRISPR-Cas9 screens using naïve and AZD1775-tolerant NCI-H82 cells; AZD1775 (0.4 μM) vs. DMSO on day 21. Each screen was conducted in two replicates. Key genes in the GCN2 pathway are highlighted (EIF2AK4 = GCN2, GCN1L1 = GCN1, PPP1R15B = CreP). Fold change (log2) < −0.5 or > 0.5, ANOVA: p < 0.05 (MEMcrispR package).
(C) Fold changes of gene representations between naïve and AZD1775-tolerant NCI-H82 cells.
(D) GO enrichment terms for under- and over-represented gene hits.
(E) Representation of selected genes across the cell cycle, amino acid-sensing, mTOR, and DNA damage pathways from the CRISPR-Cas9 screens with NCI-H82 cells.
(F) Competition assay between knockout cells and control NCI-H82 cells. The ratio was calculated by the number of GFP+ and mCherry+ cells identified by flow cytometry. Two separate sgRNAs were used per gene (n = 4). t test: *p = 0.05–0.01, **p = 0.01–0.001, ***p < 0.001.
(G) GCN2 expression in NCI-H82 WT and NCI-H82 GCN2−/− cells measured by immunoassay. HSP90 is a loading control.
(H) Effect of CDK1 phosphorylation (at tyrosine 15) upon AZD1775 treatment in GCN2−/− cells (n = 1) measured by immunoassay. HSP90 is a loading control.
(I) AlamarBlue cell viability assays with wild-type and GCN2−/− NCI-H82 cells in a titration of AZD1775. n = 6 per condition, standard error of the mean (SEM). Treatment: 5 days.
